
Granulosicoccus antarcticus IMCC3135
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Granulosicoccaceae; Granulosicoccus; Granulosicoccus antarcticus
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
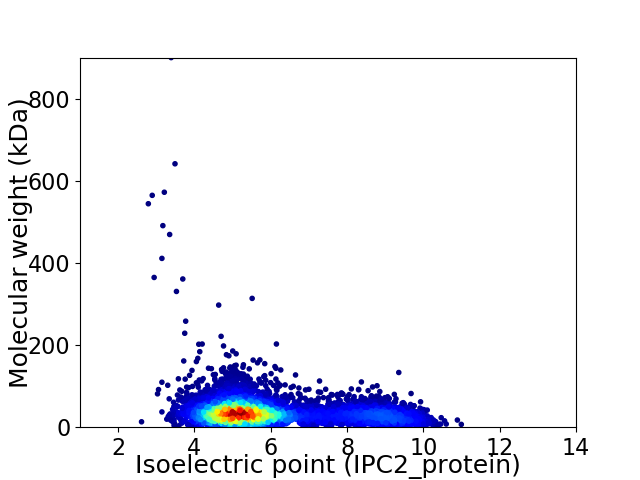
Virtual 2D-PAGE plot for 6680 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z2NMS6|A0A2Z2NMS6_9GAMM Fe2OG dioxygenase domain-containing protein OS=Granulosicoccus antarcticus IMCC3135 OX=1192854 GN=IMCC3135_12175 PE=3 SV=1
MM1 pKa = 7.49WRR3 pKa = 11.84RR4 pKa = 11.84ASVFAVHH11 pKa = 6.76IAILGSLTACSEE23 pKa = 4.58SPFSGSEE30 pKa = 3.92TADD33 pKa = 3.25SDD35 pKa = 4.22TQTSSATPSSPTSQTNDD52 pKa = 2.94GSDD55 pKa = 3.79SLSQPQSVVVAQANPTVVEE74 pKa = 4.32IPDD77 pKa = 3.4VAAATPEE84 pKa = 3.83PTPIVVNPFTPVPQPTSEE102 pKa = 4.04PDD104 pKa = 3.14EE105 pKa = 4.4EE106 pKa = 4.64QEE108 pKa = 5.03SEE110 pKa = 4.3PDD112 pKa = 3.52DD113 pKa = 4.87SPDD116 pKa = 3.32EE117 pKa = 4.19VVANSDD123 pKa = 3.41SDD125 pKa = 3.81TDD127 pKa = 3.93GNTGSDD133 pKa = 3.12AGADD137 pKa = 3.76SEE139 pKa = 5.71AEE141 pKa = 4.51LDD143 pKa = 3.84TADD146 pKa = 4.24NSPEE150 pKa = 4.02STEE153 pKa = 4.07TPEE156 pKa = 3.92TGVADD161 pKa = 4.09SQSSPTDD168 pKa = 3.26NGDD171 pKa = 3.12TDD173 pKa = 4.16LAEE176 pKa = 4.31TDD178 pKa = 3.8VDD180 pKa = 4.46DD181 pKa = 5.02SDD183 pKa = 4.71SAATDD188 pKa = 3.84TEE190 pKa = 4.33ASTDD194 pKa = 4.08DD195 pKa = 3.62NTQDD199 pKa = 5.16AEE201 pKa = 4.52TTDD204 pKa = 4.15DD205 pKa = 4.08SAPEE209 pKa = 3.67TDD211 pKa = 3.49VAEE214 pKa = 4.46EE215 pKa = 4.34SSDD218 pKa = 3.47DD219 pKa = 3.72TAEE222 pKa = 3.9QDD224 pKa = 3.61STDD227 pKa = 4.17SIVAIDD233 pKa = 4.1PADD236 pKa = 3.69SGEE239 pKa = 4.31TEE241 pKa = 3.9QSKK244 pKa = 11.4AEE246 pKa = 3.99AEE248 pKa = 4.48NNFSSHH254 pKa = 4.34TTAGFIGKK262 pKa = 9.03IVDD265 pKa = 4.47DD266 pKa = 4.9SIQVSWEE273 pKa = 4.0VEE275 pKa = 4.14PGALGYY281 pKa = 10.22NVYY284 pKa = 10.98LEE286 pKa = 4.39GKK288 pKa = 9.58YY289 pKa = 7.97VTTVWEE295 pKa = 3.95NSYY298 pKa = 11.27VEE300 pKa = 4.07TDD302 pKa = 4.47LYY304 pKa = 10.37DD305 pKa = 3.18QNYY308 pKa = 8.93YY309 pKa = 10.88YY310 pKa = 10.18EE311 pKa = 4.28IQSFDD316 pKa = 3.74EE317 pKa = 4.18TKK319 pKa = 10.49KK320 pKa = 10.28IYY322 pKa = 9.42WYY324 pKa = 10.32NATGLTVKK332 pKa = 9.9VRR334 pKa = 11.84SAGRR338 pKa = 11.84VDD340 pKa = 3.61SSRR343 pKa = 11.84ATAKK347 pKa = 10.72ADD349 pKa = 4.12LLNDD353 pKa = 3.47YY354 pKa = 10.51QLIFSDD360 pKa = 4.22EE361 pKa = 4.41FTGSTLDD368 pKa = 3.38TSKK371 pKa = 10.87WNTALLWGDD380 pKa = 3.38QIIINGEE387 pKa = 3.79VQFYY391 pKa = 11.35VDD393 pKa = 4.61INNKK397 pKa = 9.28PDD399 pKa = 3.37FGFNPFTFDD408 pKa = 5.02GEE410 pKa = 4.42HH411 pKa = 5.86LTINSIKK418 pKa = 9.65TPSEE422 pKa = 3.74LSEE425 pKa = 4.12KK426 pKa = 10.97AKK428 pKa = 10.1GQPYY432 pKa = 10.16LSGVITSYY440 pKa = 11.34DD441 pKa = 3.5ALQFIYY447 pKa = 10.05GYY449 pKa = 10.99AEE451 pKa = 3.7TRR453 pKa = 11.84AKK455 pKa = 9.38VTFGRR460 pKa = 11.84GYY462 pKa = 10.02WPAFWLLNAYY472 pKa = 9.19YY473 pKa = 10.14GQGGDD478 pKa = 4.33DD479 pKa = 3.81PEE481 pKa = 5.43IDD483 pKa = 2.86IMEE486 pKa = 5.4FIGHH490 pKa = 6.37DD491 pKa = 3.33QDD493 pKa = 4.46VVYY496 pKa = 8.4HH497 pKa = 6.4TYY499 pKa = 10.3HH500 pKa = 6.79YY501 pKa = 10.63YY502 pKa = 10.68DD503 pKa = 3.49EE504 pKa = 4.86NGEE507 pKa = 4.06LRR509 pKa = 11.84STKK512 pKa = 9.89SQPTPGVDD520 pKa = 3.39YY521 pKa = 11.56SEE523 pKa = 5.45DD524 pKa = 3.1FHH526 pKa = 7.47TFGVEE531 pKa = 3.92WKK533 pKa = 9.11PGTIIYY539 pKa = 10.0YY540 pKa = 8.9VDD542 pKa = 4.32GIEE545 pKa = 3.92VHH547 pKa = 6.52RR548 pKa = 11.84VVDD551 pKa = 4.12PKK553 pKa = 11.14VSQQTMYY560 pKa = 11.01VIANTAMGGWWAGDD574 pKa = 3.65PDD576 pKa = 4.3EE577 pKa = 4.63TTPFPGEE584 pKa = 3.69FMIDD588 pKa = 3.63YY589 pKa = 10.03IRR591 pKa = 11.84VYY593 pKa = 10.74QRR595 pKa = 11.84TTPYY599 pKa = 11.32DD600 pKa = 3.5DD601 pKa = 4.7VLFNDD606 pKa = 5.03DD607 pKa = 3.18MTGIPFADD615 pKa = 5.44EE616 pKa = 4.46IPGSAIPNHH625 pKa = 6.77RR626 pKa = 11.84PTPAQWPAGYY636 pKa = 8.83PHH638 pKa = 6.97AQQ640 pKa = 3.11
MM1 pKa = 7.49WRR3 pKa = 11.84RR4 pKa = 11.84ASVFAVHH11 pKa = 6.76IAILGSLTACSEE23 pKa = 4.58SPFSGSEE30 pKa = 3.92TADD33 pKa = 3.25SDD35 pKa = 4.22TQTSSATPSSPTSQTNDD52 pKa = 2.94GSDD55 pKa = 3.79SLSQPQSVVVAQANPTVVEE74 pKa = 4.32IPDD77 pKa = 3.4VAAATPEE84 pKa = 3.83PTPIVVNPFTPVPQPTSEE102 pKa = 4.04PDD104 pKa = 3.14EE105 pKa = 4.4EE106 pKa = 4.64QEE108 pKa = 5.03SEE110 pKa = 4.3PDD112 pKa = 3.52DD113 pKa = 4.87SPDD116 pKa = 3.32EE117 pKa = 4.19VVANSDD123 pKa = 3.41SDD125 pKa = 3.81TDD127 pKa = 3.93GNTGSDD133 pKa = 3.12AGADD137 pKa = 3.76SEE139 pKa = 5.71AEE141 pKa = 4.51LDD143 pKa = 3.84TADD146 pKa = 4.24NSPEE150 pKa = 4.02STEE153 pKa = 4.07TPEE156 pKa = 3.92TGVADD161 pKa = 4.09SQSSPTDD168 pKa = 3.26NGDD171 pKa = 3.12TDD173 pKa = 4.16LAEE176 pKa = 4.31TDD178 pKa = 3.8VDD180 pKa = 4.46DD181 pKa = 5.02SDD183 pKa = 4.71SAATDD188 pKa = 3.84TEE190 pKa = 4.33ASTDD194 pKa = 4.08DD195 pKa = 3.62NTQDD199 pKa = 5.16AEE201 pKa = 4.52TTDD204 pKa = 4.15DD205 pKa = 4.08SAPEE209 pKa = 3.67TDD211 pKa = 3.49VAEE214 pKa = 4.46EE215 pKa = 4.34SSDD218 pKa = 3.47DD219 pKa = 3.72TAEE222 pKa = 3.9QDD224 pKa = 3.61STDD227 pKa = 4.17SIVAIDD233 pKa = 4.1PADD236 pKa = 3.69SGEE239 pKa = 4.31TEE241 pKa = 3.9QSKK244 pKa = 11.4AEE246 pKa = 3.99AEE248 pKa = 4.48NNFSSHH254 pKa = 4.34TTAGFIGKK262 pKa = 9.03IVDD265 pKa = 4.47DD266 pKa = 4.9SIQVSWEE273 pKa = 4.0VEE275 pKa = 4.14PGALGYY281 pKa = 10.22NVYY284 pKa = 10.98LEE286 pKa = 4.39GKK288 pKa = 9.58YY289 pKa = 7.97VTTVWEE295 pKa = 3.95NSYY298 pKa = 11.27VEE300 pKa = 4.07TDD302 pKa = 4.47LYY304 pKa = 10.37DD305 pKa = 3.18QNYY308 pKa = 8.93YY309 pKa = 10.88YY310 pKa = 10.18EE311 pKa = 4.28IQSFDD316 pKa = 3.74EE317 pKa = 4.18TKK319 pKa = 10.49KK320 pKa = 10.28IYY322 pKa = 9.42WYY324 pKa = 10.32NATGLTVKK332 pKa = 9.9VRR334 pKa = 11.84SAGRR338 pKa = 11.84VDD340 pKa = 3.61SSRR343 pKa = 11.84ATAKK347 pKa = 10.72ADD349 pKa = 4.12LLNDD353 pKa = 3.47YY354 pKa = 10.51QLIFSDD360 pKa = 4.22EE361 pKa = 4.41FTGSTLDD368 pKa = 3.38TSKK371 pKa = 10.87WNTALLWGDD380 pKa = 3.38QIIINGEE387 pKa = 3.79VQFYY391 pKa = 11.35VDD393 pKa = 4.61INNKK397 pKa = 9.28PDD399 pKa = 3.37FGFNPFTFDD408 pKa = 5.02GEE410 pKa = 4.42HH411 pKa = 5.86LTINSIKK418 pKa = 9.65TPSEE422 pKa = 3.74LSEE425 pKa = 4.12KK426 pKa = 10.97AKK428 pKa = 10.1GQPYY432 pKa = 10.16LSGVITSYY440 pKa = 11.34DD441 pKa = 3.5ALQFIYY447 pKa = 10.05GYY449 pKa = 10.99AEE451 pKa = 3.7TRR453 pKa = 11.84AKK455 pKa = 9.38VTFGRR460 pKa = 11.84GYY462 pKa = 10.02WPAFWLLNAYY472 pKa = 9.19YY473 pKa = 10.14GQGGDD478 pKa = 4.33DD479 pKa = 3.81PEE481 pKa = 5.43IDD483 pKa = 2.86IMEE486 pKa = 5.4FIGHH490 pKa = 6.37DD491 pKa = 3.33QDD493 pKa = 4.46VVYY496 pKa = 8.4HH497 pKa = 6.4TYY499 pKa = 10.3HH500 pKa = 6.79YY501 pKa = 10.63YY502 pKa = 10.68DD503 pKa = 3.49EE504 pKa = 4.86NGEE507 pKa = 4.06LRR509 pKa = 11.84STKK512 pKa = 9.89SQPTPGVDD520 pKa = 3.39YY521 pKa = 11.56SEE523 pKa = 5.45DD524 pKa = 3.1FHH526 pKa = 7.47TFGVEE531 pKa = 3.92WKK533 pKa = 9.11PGTIIYY539 pKa = 10.0YY540 pKa = 8.9VDD542 pKa = 4.32GIEE545 pKa = 3.92VHH547 pKa = 6.52RR548 pKa = 11.84VVDD551 pKa = 4.12PKK553 pKa = 11.14VSQQTMYY560 pKa = 11.01VIANTAMGGWWAGDD574 pKa = 3.65PDD576 pKa = 4.3EE577 pKa = 4.63TTPFPGEE584 pKa = 3.69FMIDD588 pKa = 3.63YY589 pKa = 10.03IRR591 pKa = 11.84VYY593 pKa = 10.74QRR595 pKa = 11.84TTPYY599 pKa = 11.32DD600 pKa = 3.5DD601 pKa = 4.7VLFNDD606 pKa = 5.03DD607 pKa = 3.18MTGIPFADD615 pKa = 5.44EE616 pKa = 4.46IPGSAIPNHH625 pKa = 6.77RR626 pKa = 11.84PTPAQWPAGYY636 pKa = 8.83PHH638 pKa = 6.97AQQ640 pKa = 3.11
Molecular weight: 69.97 kDa
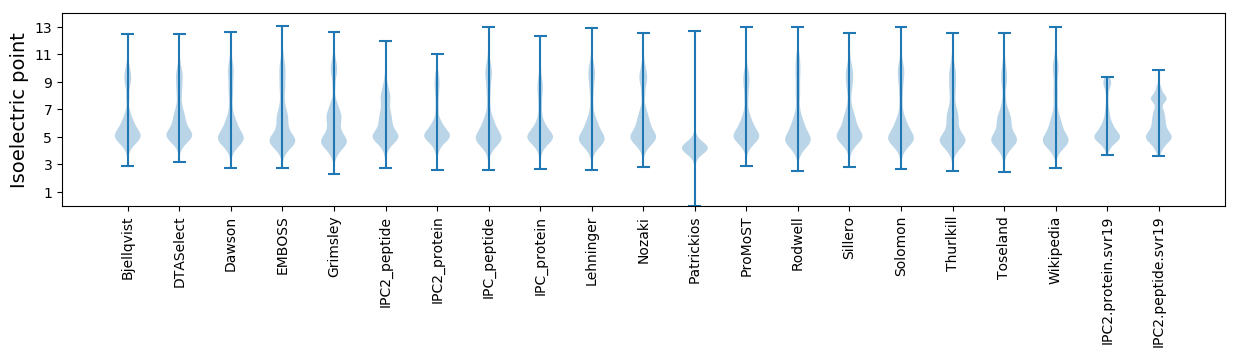
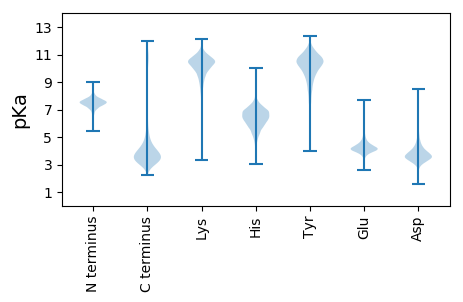
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z2NWN7|A0A2Z2NWN7_9GAMM 8-oxo-dGTP diphosphatase OS=Granulosicoccus antarcticus IMCC3135 OX=1192854 GN=mutT PE=3 SV=1
MM1 pKa = 7.53INSIPPQGLVLLSIVAIQVGAAVAIHH27 pKa = 6.57LFPVLGASGTVAVRR41 pKa = 11.84IIFSAILLGLAARR54 pKa = 11.84TGLRR58 pKa = 11.84TLVQTFRR65 pKa = 11.84LHH67 pKa = 5.41WKK69 pKa = 9.4LLVVFGLSIAAMNLFFYY86 pKa = 10.42QALARR91 pKa = 11.84IPLGAAVAFEE101 pKa = 4.69FIGPLGVAALASRR114 pKa = 11.84RR115 pKa = 11.84LTQFAWIALAALGIVLLSPLSGADD139 pKa = 3.77LDD141 pKa = 4.8PIGILFALMAGTGWACFIIFAGRR164 pKa = 11.84VGKK167 pKa = 9.91LIPGNDD173 pKa = 3.4GLAIGMAVAALAMIPFALPVAGILVTNPLILLASIGVALLSTTIPFTFEE222 pKa = 3.19FTALKK227 pKa = 10.11RR228 pKa = 11.84LPARR232 pKa = 11.84TYY234 pKa = 9.98GVLVSLEE241 pKa = 4.23PGVAALVGALLLGEE255 pKa = 4.82RR256 pKa = 11.84IGLQGMIAITCVIIAAIGITLSDD279 pKa = 3.64ARR281 pKa = 11.84TPPP284 pKa = 3.23
MM1 pKa = 7.53INSIPPQGLVLLSIVAIQVGAAVAIHH27 pKa = 6.57LFPVLGASGTVAVRR41 pKa = 11.84IIFSAILLGLAARR54 pKa = 11.84TGLRR58 pKa = 11.84TLVQTFRR65 pKa = 11.84LHH67 pKa = 5.41WKK69 pKa = 9.4LLVVFGLSIAAMNLFFYY86 pKa = 10.42QALARR91 pKa = 11.84IPLGAAVAFEE101 pKa = 4.69FIGPLGVAALASRR114 pKa = 11.84RR115 pKa = 11.84LTQFAWIALAALGIVLLSPLSGADD139 pKa = 3.77LDD141 pKa = 4.8PIGILFALMAGTGWACFIIFAGRR164 pKa = 11.84VGKK167 pKa = 9.91LIPGNDD173 pKa = 3.4GLAIGMAVAALAMIPFALPVAGILVTNPLILLASIGVALLSTTIPFTFEE222 pKa = 3.19FTALKK227 pKa = 10.11RR228 pKa = 11.84LPARR232 pKa = 11.84TYY234 pKa = 9.98GVLVSLEE241 pKa = 4.23PGVAALVGALLLGEE255 pKa = 4.82RR256 pKa = 11.84IGLQGMIAITCVIIAAIGITLSDD279 pKa = 3.64ARR281 pKa = 11.84TPPP284 pKa = 3.23
Molecular weight: 29.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2309472 |
29 |
8704 |
345.7 |
37.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.024 ± 0.032 | 1.035 ± 0.013 |
6.086 ± 0.077 | 5.862 ± 0.028 |
3.704 ± 0.02 | 7.601 ± 0.031 |
2.137 ± 0.018 | 5.678 ± 0.025 |
3.376 ± 0.033 | 10.503 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.528 ± 0.022 | 3.47 ± 0.031 |
4.434 ± 0.024 | 4.013 ± 0.026 |
5.665 ± 0.044 | 7.147 ± 0.038 |
5.656 ± 0.054 | 7.271 ± 0.027 |
1.304 ± 0.014 | 2.507 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
