
Flavobacterium phage vB_FspS_tooticki6-1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Muminvirus; Flavobacterium virus Tooticki
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
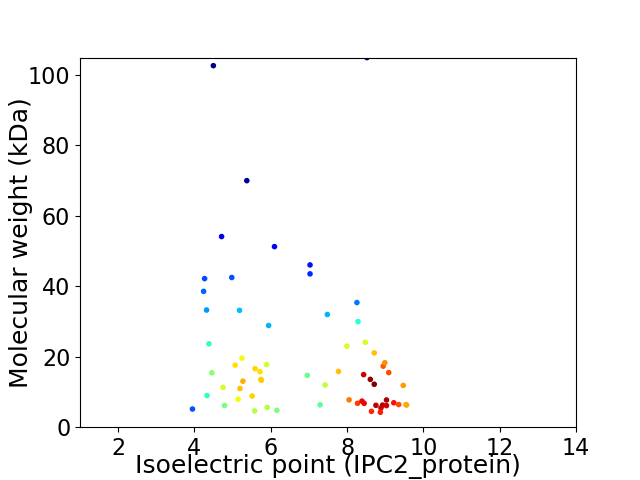
Virtual 2D-PAGE plot for 65 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6B9LPE1|A0A6B9LPE1_9CAUD DNA methyltransferase OS=Flavobacterium phage vB_FspS_tooticki6-1 OX=2686279 GN=tooticki61_gp005 PE=4 SV=1
MM1 pKa = 7.67NDD3 pKa = 3.11SIIRR7 pKa = 11.84FKK9 pKa = 11.46NSLNDD14 pKa = 4.29ALNLDD19 pKa = 4.43LNDD22 pKa = 3.76NAILGYY28 pKa = 10.96ANQIVLNPFKK38 pKa = 10.48FYY40 pKa = 8.43TQKK43 pKa = 9.79TPIEE47 pKa = 4.21MIFEE51 pKa = 4.37GEE53 pKa = 4.24YY54 pKa = 9.23TCYY57 pKa = 10.54ISDD60 pKa = 3.39ICGDD64 pKa = 3.7NLQDD68 pKa = 3.64ITEE71 pKa = 4.07HH72 pKa = 6.25TYY74 pKa = 10.49ISEE77 pKa = 4.18NTNGNYY83 pKa = 9.4IEE85 pKa = 4.65FATGVDD91 pKa = 4.24FQRR94 pKa = 11.84KK95 pKa = 8.21LVLIKK100 pKa = 10.45LVNNINPLEE109 pKa = 3.95IWYY112 pKa = 9.41SNPMFITEE120 pKa = 4.31NVNLTTEE127 pKa = 4.42FDD129 pKa = 3.99YY130 pKa = 11.66KK131 pKa = 10.86NVSDD135 pKa = 5.88AYY137 pKa = 8.13MQSVALEE144 pKa = 4.25CFFTRR149 pKa = 11.84SIQEE153 pKa = 4.29SEE155 pKa = 4.23VKK157 pKa = 10.53SYY159 pKa = 10.39VQEE162 pKa = 4.2TGTKK166 pKa = 9.48VSGKK170 pKa = 8.87ATFIEE175 pKa = 3.93MRR177 pKa = 11.84KK178 pKa = 9.81FIFEE182 pKa = 3.94MLDD185 pKa = 3.02NFVYY189 pKa = 10.36RR190 pKa = 11.84RR191 pKa = 11.84LNLILAKK198 pKa = 9.25TQLYY202 pKa = 8.12VQEE205 pKa = 4.43IRR207 pKa = 11.84VTDD210 pKa = 3.75KK211 pKa = 11.14PLLKK215 pKa = 10.87DD216 pKa = 3.68SDD218 pKa = 3.6IQGNQNTFSSEE229 pKa = 3.45FTGAVNYY236 pKa = 10.62NDD238 pKa = 3.69TYY240 pKa = 11.42TRR242 pKa = 11.84TLQIAQPLALVSNYY256 pKa = 9.77PNAIYY261 pKa = 9.83TLDD264 pKa = 4.15TISDD268 pKa = 5.03LIQLVFNHH276 pKa = 7.07DD277 pKa = 3.33VDD279 pKa = 4.02TDD281 pKa = 3.93NEE283 pKa = 4.13DD284 pKa = 3.77LQISLYY290 pKa = 10.02KK291 pKa = 10.7DD292 pKa = 3.56EE293 pKa = 5.43IFISYY298 pKa = 10.65LDD300 pKa = 4.17LIKK303 pKa = 10.69INFTTFEE310 pKa = 3.74QVYY313 pKa = 9.26NFVSNGEE320 pKa = 4.02YY321 pKa = 10.44KK322 pKa = 10.37IIIPANKK329 pKa = 9.46YY330 pKa = 10.6SSILYY335 pKa = 10.22GSLPYY340 pKa = 10.25TEE342 pKa = 4.73LTFTILGGEE351 pKa = 4.35YY352 pKa = 10.64EE353 pKa = 4.11NTEE356 pKa = 4.14YY357 pKa = 11.2NDD359 pKa = 3.59EE360 pKa = 4.16YY361 pKa = 11.52LLNN364 pKa = 4.17
MM1 pKa = 7.67NDD3 pKa = 3.11SIIRR7 pKa = 11.84FKK9 pKa = 11.46NSLNDD14 pKa = 4.29ALNLDD19 pKa = 4.43LNDD22 pKa = 3.76NAILGYY28 pKa = 10.96ANQIVLNPFKK38 pKa = 10.48FYY40 pKa = 8.43TQKK43 pKa = 9.79TPIEE47 pKa = 4.21MIFEE51 pKa = 4.37GEE53 pKa = 4.24YY54 pKa = 9.23TCYY57 pKa = 10.54ISDD60 pKa = 3.39ICGDD64 pKa = 3.7NLQDD68 pKa = 3.64ITEE71 pKa = 4.07HH72 pKa = 6.25TYY74 pKa = 10.49ISEE77 pKa = 4.18NTNGNYY83 pKa = 9.4IEE85 pKa = 4.65FATGVDD91 pKa = 4.24FQRR94 pKa = 11.84KK95 pKa = 8.21LVLIKK100 pKa = 10.45LVNNINPLEE109 pKa = 3.95IWYY112 pKa = 9.41SNPMFITEE120 pKa = 4.31NVNLTTEE127 pKa = 4.42FDD129 pKa = 3.99YY130 pKa = 11.66KK131 pKa = 10.86NVSDD135 pKa = 5.88AYY137 pKa = 8.13MQSVALEE144 pKa = 4.25CFFTRR149 pKa = 11.84SIQEE153 pKa = 4.29SEE155 pKa = 4.23VKK157 pKa = 10.53SYY159 pKa = 10.39VQEE162 pKa = 4.2TGTKK166 pKa = 9.48VSGKK170 pKa = 8.87ATFIEE175 pKa = 3.93MRR177 pKa = 11.84KK178 pKa = 9.81FIFEE182 pKa = 3.94MLDD185 pKa = 3.02NFVYY189 pKa = 10.36RR190 pKa = 11.84RR191 pKa = 11.84LNLILAKK198 pKa = 9.25TQLYY202 pKa = 8.12VQEE205 pKa = 4.43IRR207 pKa = 11.84VTDD210 pKa = 3.75KK211 pKa = 11.14PLLKK215 pKa = 10.87DD216 pKa = 3.68SDD218 pKa = 3.6IQGNQNTFSSEE229 pKa = 3.45FTGAVNYY236 pKa = 10.62NDD238 pKa = 3.69TYY240 pKa = 11.42TRR242 pKa = 11.84TLQIAQPLALVSNYY256 pKa = 9.77PNAIYY261 pKa = 9.83TLDD264 pKa = 4.15TISDD268 pKa = 5.03LIQLVFNHH276 pKa = 7.07DD277 pKa = 3.33VDD279 pKa = 4.02TDD281 pKa = 3.93NEE283 pKa = 4.13DD284 pKa = 3.77LQISLYY290 pKa = 10.02KK291 pKa = 10.7DD292 pKa = 3.56EE293 pKa = 5.43IFISYY298 pKa = 10.65LDD300 pKa = 4.17LIKK303 pKa = 10.69INFTTFEE310 pKa = 3.74QVYY313 pKa = 9.26NFVSNGEE320 pKa = 4.02YY321 pKa = 10.44KK322 pKa = 10.37IIIPANKK329 pKa = 9.46YY330 pKa = 10.6SSILYY335 pKa = 10.22GSLPYY340 pKa = 10.25TEE342 pKa = 4.73LTFTILGGEE351 pKa = 4.35YY352 pKa = 10.64EE353 pKa = 4.11NTEE356 pKa = 4.14YY357 pKa = 11.2NDD359 pKa = 3.59EE360 pKa = 4.16YY361 pKa = 11.52LLNN364 pKa = 4.17
Molecular weight: 42.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6B9LP10|A0A6B9LP10_9CAUD Uncharacterized protein OS=Flavobacterium phage vB_FspS_tooticki6-1 OX=2686279 GN=tooticki61_gp038 PE=4 SV=1
MM1 pKa = 7.51SKK3 pKa = 10.45LSKK6 pKa = 10.2RR7 pKa = 11.84KK8 pKa = 10.25SPVLVGNRR16 pKa = 11.84IDD18 pKa = 3.47TTYY21 pKa = 10.81EE22 pKa = 3.51RR23 pKa = 11.84QRR25 pKa = 11.84NITQTAKK32 pKa = 10.36EE33 pKa = 4.08VAEE36 pKa = 4.23RR37 pKa = 11.84TSEE40 pKa = 4.08EE41 pKa = 3.98IKK43 pKa = 10.91NKK45 pKa = 10.09NIRR48 pKa = 11.84YY49 pKa = 9.08DD50 pKa = 3.14IKK52 pKa = 10.88RR53 pKa = 3.47
MM1 pKa = 7.51SKK3 pKa = 10.45LSKK6 pKa = 10.2RR7 pKa = 11.84KK8 pKa = 10.25SPVLVGNRR16 pKa = 11.84IDD18 pKa = 3.47TTYY21 pKa = 10.81EE22 pKa = 3.51RR23 pKa = 11.84QRR25 pKa = 11.84NITQTAKK32 pKa = 10.36EE33 pKa = 4.08VAEE36 pKa = 4.23RR37 pKa = 11.84TSEE40 pKa = 4.08EE41 pKa = 3.98IKK43 pKa = 10.91NKK45 pKa = 10.09NIRR48 pKa = 11.84YY49 pKa = 9.08DD50 pKa = 3.14IKK52 pKa = 10.88RR53 pKa = 3.47
Molecular weight: 6.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11722 |
35 |
930 |
180.3 |
20.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.905 ± 0.35 | 1.177 ± 0.185 |
5.648 ± 0.279 | 7.166 ± 0.321 |
5.255 ± 0.244 | 4.718 ± 0.248 |
1.322 ± 0.178 | 8.045 ± 0.281 |
9.469 ± 0.571 | 8.966 ± 0.294 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.124 ± 0.122 | 8.164 ± 0.316 |
2.423 ± 0.193 | 3.924 ± 0.318 |
3.046 ± 0.184 | 6.501 ± 0.276 |
6.637 ± 0.367 | 5.332 ± 0.207 |
0.896 ± 0.079 | 4.283 ± 0.237 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
