
Marinobacter sp. F3R11
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Marinobacter; unclassified Marinobacter
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
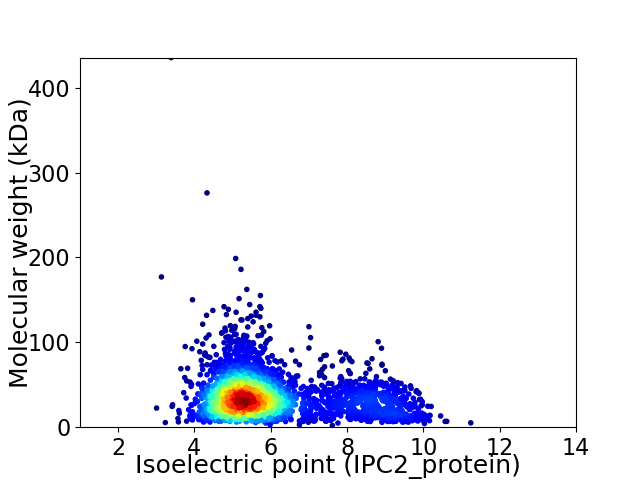
Virtual 2D-PAGE plot for 3017 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
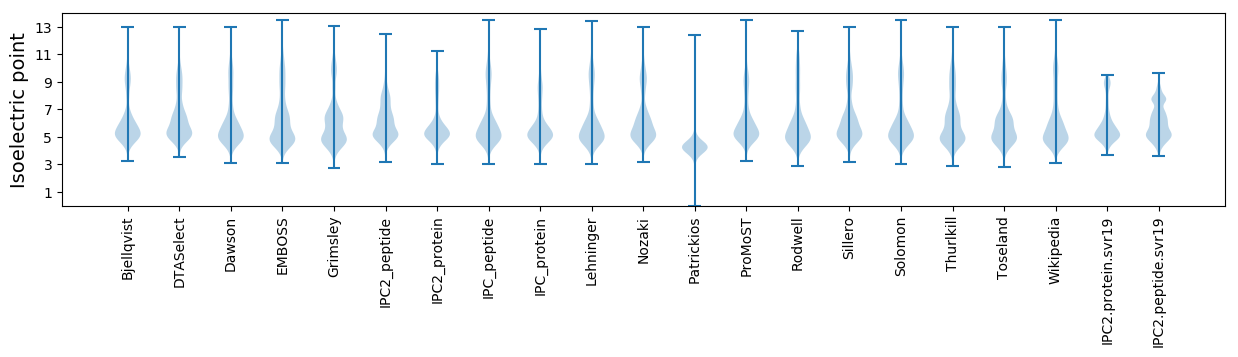
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A366WJ66|A0A366WJ66_9ALTE Flagellar motor protein MotD OS=Marinobacter sp. F3R11 OX=2267231 GN=DS878_06825 PE=3 SV=1
MM1 pKa = 7.31AQQHH5 pKa = 5.71KK6 pKa = 10.74LKK8 pKa = 10.64FLVPALGLMMTACGGSDD25 pKa = 3.16NKK27 pKa = 10.33ISEE30 pKa = 4.15QADD33 pKa = 3.36DD34 pKa = 5.51GGASDD39 pKa = 4.8NDD41 pKa = 3.58YY42 pKa = 11.1SQLVIDD48 pKa = 5.58ARR50 pKa = 11.84TQTQYY55 pKa = 12.02LNLATGEE62 pKa = 4.45VVSSDD67 pKa = 3.93GDD69 pKa = 3.07WDD71 pKa = 3.86LAFNRR76 pKa = 11.84TSVLLNSGASGNGNVAGAMGDD97 pKa = 3.79EE98 pKa = 4.04QAEE101 pKa = 4.49FYY103 pKa = 11.02DD104 pKa = 4.56ADD106 pKa = 4.33GNPDD110 pKa = 3.29LNKK113 pKa = 10.06FVGATADD120 pKa = 3.86GEE122 pKa = 4.34LDD124 pKa = 3.42HH125 pKa = 7.14LKK127 pKa = 10.93DD128 pKa = 3.68SFPAPEE134 pKa = 4.03SWISDD139 pKa = 3.39DD140 pKa = 3.4VVYY143 pKa = 11.09AFGDD147 pKa = 3.45GWSVYY152 pKa = 10.79GDD154 pKa = 3.19GGVISEE160 pKa = 4.59VPDD163 pKa = 2.76IGYY166 pKa = 9.74LVRR169 pKa = 11.84SAEE172 pKa = 4.04GDD174 pKa = 3.25SYY176 pKa = 12.0ARR178 pKa = 11.84MRR180 pKa = 11.84IVDD183 pKa = 3.89FNFPTRR189 pKa = 11.84SGNGIEE195 pKa = 4.22SFNLEE200 pKa = 4.42FEE202 pKa = 4.51VQSAGTTQLSGTTINFTEE220 pKa = 4.25PLEE223 pKa = 4.17YY224 pKa = 10.61DD225 pKa = 3.58GGDD228 pKa = 3.02ACFDD232 pKa = 3.85FDD234 pKa = 4.02TNAVVDD240 pKa = 4.72CSSSDD245 pKa = 3.05TWDD248 pKa = 3.33VLIGFSGRR256 pKa = 11.84EE257 pKa = 3.87WYY259 pKa = 10.81LKK261 pKa = 9.69TNSGPSGAGQGGALGPIDD279 pKa = 3.8WSEE282 pKa = 3.77LSAYY286 pKa = 9.13TSATIDD292 pKa = 3.54SATGEE297 pKa = 4.56SLTQAYY303 pKa = 10.17ASDD306 pKa = 3.73STGGLFTDD314 pKa = 4.17NSWYY318 pKa = 10.52AYY320 pKa = 10.19NLQGAHH326 pKa = 6.89KK327 pKa = 9.12LWPNFRR333 pKa = 11.84VYY335 pKa = 10.97LIDD338 pKa = 4.67SDD340 pKa = 5.39SEE342 pKa = 4.16DD343 pKa = 3.3ASAPVYY349 pKa = 10.96AMQIINYY356 pKa = 8.84YY357 pKa = 9.78GADD360 pKa = 3.78GNSGQPEE367 pKa = 4.16IRR369 pKa = 11.84WKK371 pKa = 10.52EE372 pKa = 3.6ISLTTGEE379 pKa = 4.11NN380 pKa = 3.21
MM1 pKa = 7.31AQQHH5 pKa = 5.71KK6 pKa = 10.74LKK8 pKa = 10.64FLVPALGLMMTACGGSDD25 pKa = 3.16NKK27 pKa = 10.33ISEE30 pKa = 4.15QADD33 pKa = 3.36DD34 pKa = 5.51GGASDD39 pKa = 4.8NDD41 pKa = 3.58YY42 pKa = 11.1SQLVIDD48 pKa = 5.58ARR50 pKa = 11.84TQTQYY55 pKa = 12.02LNLATGEE62 pKa = 4.45VVSSDD67 pKa = 3.93GDD69 pKa = 3.07WDD71 pKa = 3.86LAFNRR76 pKa = 11.84TSVLLNSGASGNGNVAGAMGDD97 pKa = 3.79EE98 pKa = 4.04QAEE101 pKa = 4.49FYY103 pKa = 11.02DD104 pKa = 4.56ADD106 pKa = 4.33GNPDD110 pKa = 3.29LNKK113 pKa = 10.06FVGATADD120 pKa = 3.86GEE122 pKa = 4.34LDD124 pKa = 3.42HH125 pKa = 7.14LKK127 pKa = 10.93DD128 pKa = 3.68SFPAPEE134 pKa = 4.03SWISDD139 pKa = 3.39DD140 pKa = 3.4VVYY143 pKa = 11.09AFGDD147 pKa = 3.45GWSVYY152 pKa = 10.79GDD154 pKa = 3.19GGVISEE160 pKa = 4.59VPDD163 pKa = 2.76IGYY166 pKa = 9.74LVRR169 pKa = 11.84SAEE172 pKa = 4.04GDD174 pKa = 3.25SYY176 pKa = 12.0ARR178 pKa = 11.84MRR180 pKa = 11.84IVDD183 pKa = 3.89FNFPTRR189 pKa = 11.84SGNGIEE195 pKa = 4.22SFNLEE200 pKa = 4.42FEE202 pKa = 4.51VQSAGTTQLSGTTINFTEE220 pKa = 4.25PLEE223 pKa = 4.17YY224 pKa = 10.61DD225 pKa = 3.58GGDD228 pKa = 3.02ACFDD232 pKa = 3.85FDD234 pKa = 4.02TNAVVDD240 pKa = 4.72CSSSDD245 pKa = 3.05TWDD248 pKa = 3.33VLIGFSGRR256 pKa = 11.84EE257 pKa = 3.87WYY259 pKa = 10.81LKK261 pKa = 9.69TNSGPSGAGQGGALGPIDD279 pKa = 3.8WSEE282 pKa = 3.77LSAYY286 pKa = 9.13TSATIDD292 pKa = 3.54SATGEE297 pKa = 4.56SLTQAYY303 pKa = 10.17ASDD306 pKa = 3.73STGGLFTDD314 pKa = 4.17NSWYY318 pKa = 10.52AYY320 pKa = 10.19NLQGAHH326 pKa = 6.89KK327 pKa = 9.12LWPNFRR333 pKa = 11.84VYY335 pKa = 10.97LIDD338 pKa = 4.67SDD340 pKa = 5.39SEE342 pKa = 4.16DD343 pKa = 3.3ASAPVYY349 pKa = 10.96AMQIINYY356 pKa = 8.84YY357 pKa = 9.78GADD360 pKa = 3.78GNSGQPEE367 pKa = 4.16IRR369 pKa = 11.84WKK371 pKa = 10.52EE372 pKa = 3.6ISLTTGEE379 pKa = 4.11NN380 pKa = 3.21
Molecular weight: 40.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A366WVL1|A0A366WVL1_9ALTE Bifunctional protein GlmU OS=Marinobacter sp. F3R11 OX=2267231 GN=glmU PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.34RR12 pKa = 11.84KK13 pKa = 9.1RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 9.34VISRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.41GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.34RR12 pKa = 11.84KK13 pKa = 9.1RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 9.34VISRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.41GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1025962 |
17 |
4222 |
340.1 |
37.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.694 ± 0.042 | 0.965 ± 0.016 |
5.67 ± 0.043 | 6.514 ± 0.047 |
3.804 ± 0.029 | 7.891 ± 0.041 |
2.148 ± 0.023 | 5.507 ± 0.037 |
3.775 ± 0.033 | 10.614 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.637 ± 0.024 | 3.33 ± 0.031 |
4.63 ± 0.032 | 3.907 ± 0.03 |
6.164 ± 0.036 | 6.25 ± 0.039 |
5.23 ± 0.038 | 7.309 ± 0.038 |
1.343 ± 0.02 | 2.617 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
