
Agromyces sp. CFH 90414
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Agromyces; unclassified Agromyces
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
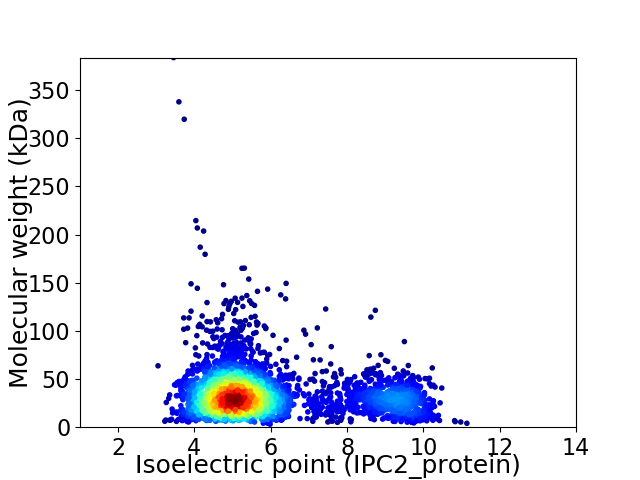
Virtual 2D-PAGE plot for 3536 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I2FFP7|A0A6I2FFP7_9MICO Alpha-mann_mid domain-containing protein OS=Agromyces sp. CFH 90414 OX=2662258 GN=GE115_12760 PE=3 SV=1
MM1 pKa = 7.12SRR3 pKa = 11.84KK4 pKa = 9.16IGRR7 pKa = 11.84IVAASLATLAVAATAGCTSAGSGDD31 pKa = 3.77SGGGTIEE38 pKa = 4.4LWTHH42 pKa = 6.2AGGNEE47 pKa = 3.94AEE49 pKa = 4.59LGVITSMVDD58 pKa = 3.4DD59 pKa = 5.38FNASQDD65 pKa = 3.75DD66 pKa = 4.15YY67 pKa = 11.03TVEE70 pKa = 4.03ITDD73 pKa = 4.57FPQDD77 pKa = 3.26AYY79 pKa = 11.67NDD81 pKa = 3.93AVVAAATSDD90 pKa = 3.77NLPCIVDD97 pKa = 3.02IDD99 pKa = 4.27GPNVANWAWAGYY111 pKa = 8.28IQPLGLPEE119 pKa = 3.99EE120 pKa = 4.73TYY122 pKa = 10.98DD123 pKa = 3.69GQLAGTLGVVDD134 pKa = 4.9DD135 pKa = 4.41EE136 pKa = 5.09VYY138 pKa = 10.56AYY140 pKa = 10.75GFFDD144 pKa = 3.21VALAMFSRR152 pKa = 11.84EE153 pKa = 3.93STLADD158 pKa = 3.15AGVRR162 pKa = 11.84VPTVDD167 pKa = 3.76EE168 pKa = 4.4PWTGEE173 pKa = 3.91EE174 pKa = 4.72FADD177 pKa = 4.2ALAKK181 pKa = 9.93IDD183 pKa = 3.4GLNRR187 pKa = 11.84FDD189 pKa = 3.95YY190 pKa = 11.12AVDD193 pKa = 4.45FGTGGGGEE201 pKa = 4.71WIPYY205 pKa = 9.58AYY207 pKa = 10.4SPLLQSFGGDD217 pKa = 3.31LIDD220 pKa = 5.4RR221 pKa = 11.84SDD223 pKa = 3.52FQTADD228 pKa = 3.19GVLNGPEE235 pKa = 3.65ALAWAEE241 pKa = 4.04WFRR244 pKa = 11.84GLVDD248 pKa = 3.52SGYY251 pKa = 8.61MAQASGEE258 pKa = 4.15DD259 pKa = 3.34ATADD263 pKa = 3.78FVNGTSGILYY273 pKa = 7.9TGNWADD279 pKa = 3.57TTVRR283 pKa = 11.84EE284 pKa = 4.34AFDD287 pKa = 4.16DD288 pKa = 4.38AVVLPPPDD296 pKa = 4.41FGDD299 pKa = 4.06GPKK302 pKa = 9.83IGAGSWQWGLTTGCDD317 pKa = 3.6DD318 pKa = 3.39VDD320 pKa = 3.59GARR323 pKa = 11.84AYY325 pKa = 11.18LEE327 pKa = 4.24FAHH330 pKa = 6.07DD331 pKa = 3.25TKK333 pKa = 11.55YY334 pKa = 10.43FVQYY338 pKa = 11.22GEE340 pKa = 4.15ATGLIPATLDD350 pKa = 3.12AAAEE354 pKa = 4.08IPAYY358 pKa = 10.47AEE360 pKa = 4.38GGDD363 pKa = 3.38NRR365 pKa = 11.84IYY367 pKa = 11.1LEE369 pKa = 4.66LADD372 pKa = 3.6QFAVVRR378 pKa = 11.84PEE380 pKa = 3.7TAAYY384 pKa = 8.6PYY386 pKa = 10.07ISSVFQKK393 pKa = 9.54ATQDD397 pKa = 3.29ILSGGDD403 pKa = 3.1AKK405 pKa = 11.17SILDD409 pKa = 3.98KK410 pKa = 11.03AVSDD414 pKa = 3.72IDD416 pKa = 5.63RR417 pKa = 11.84DD418 pKa = 3.55IQQNGGYY425 pKa = 9.83EE426 pKa = 3.98FF427 pKa = 4.93
MM1 pKa = 7.12SRR3 pKa = 11.84KK4 pKa = 9.16IGRR7 pKa = 11.84IVAASLATLAVAATAGCTSAGSGDD31 pKa = 3.77SGGGTIEE38 pKa = 4.4LWTHH42 pKa = 6.2AGGNEE47 pKa = 3.94AEE49 pKa = 4.59LGVITSMVDD58 pKa = 3.4DD59 pKa = 5.38FNASQDD65 pKa = 3.75DD66 pKa = 4.15YY67 pKa = 11.03TVEE70 pKa = 4.03ITDD73 pKa = 4.57FPQDD77 pKa = 3.26AYY79 pKa = 11.67NDD81 pKa = 3.93AVVAAATSDD90 pKa = 3.77NLPCIVDD97 pKa = 3.02IDD99 pKa = 4.27GPNVANWAWAGYY111 pKa = 8.28IQPLGLPEE119 pKa = 3.99EE120 pKa = 4.73TYY122 pKa = 10.98DD123 pKa = 3.69GQLAGTLGVVDD134 pKa = 4.9DD135 pKa = 4.41EE136 pKa = 5.09VYY138 pKa = 10.56AYY140 pKa = 10.75GFFDD144 pKa = 3.21VALAMFSRR152 pKa = 11.84EE153 pKa = 3.93STLADD158 pKa = 3.15AGVRR162 pKa = 11.84VPTVDD167 pKa = 3.76EE168 pKa = 4.4PWTGEE173 pKa = 3.91EE174 pKa = 4.72FADD177 pKa = 4.2ALAKK181 pKa = 9.93IDD183 pKa = 3.4GLNRR187 pKa = 11.84FDD189 pKa = 3.95YY190 pKa = 11.12AVDD193 pKa = 4.45FGTGGGGEE201 pKa = 4.71WIPYY205 pKa = 9.58AYY207 pKa = 10.4SPLLQSFGGDD217 pKa = 3.31LIDD220 pKa = 5.4RR221 pKa = 11.84SDD223 pKa = 3.52FQTADD228 pKa = 3.19GVLNGPEE235 pKa = 3.65ALAWAEE241 pKa = 4.04WFRR244 pKa = 11.84GLVDD248 pKa = 3.52SGYY251 pKa = 8.61MAQASGEE258 pKa = 4.15DD259 pKa = 3.34ATADD263 pKa = 3.78FVNGTSGILYY273 pKa = 7.9TGNWADD279 pKa = 3.57TTVRR283 pKa = 11.84EE284 pKa = 4.34AFDD287 pKa = 4.16DD288 pKa = 4.38AVVLPPPDD296 pKa = 4.41FGDD299 pKa = 4.06GPKK302 pKa = 9.83IGAGSWQWGLTTGCDD317 pKa = 3.6DD318 pKa = 3.39VDD320 pKa = 3.59GARR323 pKa = 11.84AYY325 pKa = 11.18LEE327 pKa = 4.24FAHH330 pKa = 6.07DD331 pKa = 3.25TKK333 pKa = 11.55YY334 pKa = 10.43FVQYY338 pKa = 11.22GEE340 pKa = 4.15ATGLIPATLDD350 pKa = 3.12AAAEE354 pKa = 4.08IPAYY358 pKa = 10.47AEE360 pKa = 4.38GGDD363 pKa = 3.38NRR365 pKa = 11.84IYY367 pKa = 11.1LEE369 pKa = 4.66LADD372 pKa = 3.6QFAVVRR378 pKa = 11.84PEE380 pKa = 3.7TAAYY384 pKa = 8.6PYY386 pKa = 10.07ISSVFQKK393 pKa = 9.54ATQDD397 pKa = 3.29ILSGGDD403 pKa = 3.1AKK405 pKa = 11.17SILDD409 pKa = 3.98KK410 pKa = 11.03AVSDD414 pKa = 3.72IDD416 pKa = 5.63RR417 pKa = 11.84DD418 pKa = 3.55IQQNGGYY425 pKa = 9.83EE426 pKa = 3.98FF427 pKa = 4.93
Molecular weight: 45.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I2FFI2|A0A6I2FFI2_9MICO Methylated-DNA--protein-cysteine methyltransferase OS=Agromyces sp. CFH 90414 OX=2662258 GN=GE115_16505 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1176171 |
29 |
3702 |
332.6 |
35.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.395 ± 0.067 | 0.465 ± 0.01 |
6.339 ± 0.046 | 5.974 ± 0.035 |
3.132 ± 0.025 | 9.315 ± 0.045 |
1.941 ± 0.021 | 4.203 ± 0.035 |
1.632 ± 0.028 | 10.009 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.616 ± 0.018 | 1.75 ± 0.02 |
5.628 ± 0.036 | 2.536 ± 0.021 |
7.67 ± 0.054 | 5.237 ± 0.026 |
5.642 ± 0.041 | 9.078 ± 0.046 |
1.532 ± 0.018 | 1.904 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
