
Lachnospiraceae bacterium MD335
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; unclassified Lachnospiraceae
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
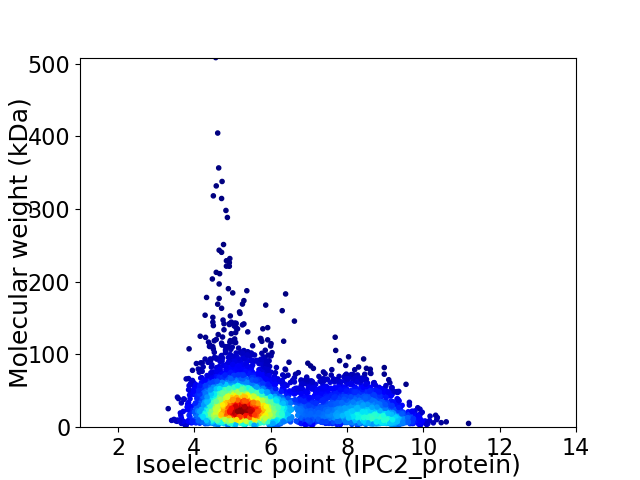
Virtual 2D-PAGE plot for 4614 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
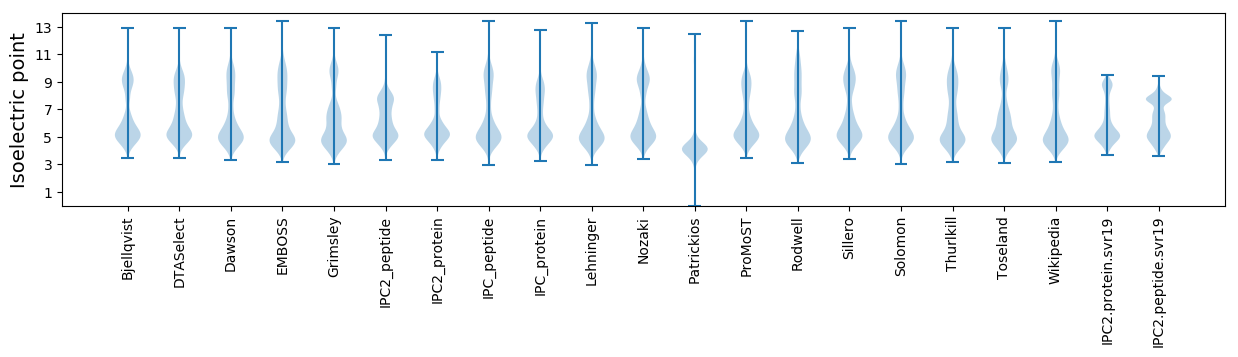
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9LBG8|R9LBG8_9FIRM Diguanylate cyclase (GGDEF) domain-containing protein OS=Lachnospiraceae bacterium MD335 OX=1235793 GN=C809_00037 PE=4 SV=1
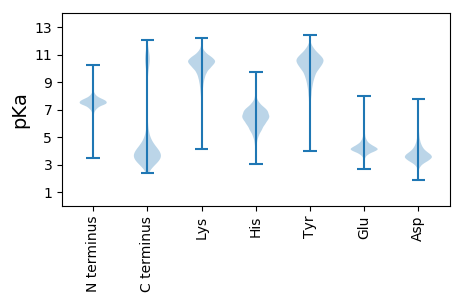
MM1 pKa = 7.84KK2 pKa = 10.01KK3 pKa = 10.32RR4 pKa = 11.84KK5 pKa = 8.19ITALLMSLVMVCSLTACGGKK25 pKa = 8.34DD26 pKa = 3.07TTTTQPEE33 pKa = 4.38PAPAPAPADD42 pKa = 3.41NAATQDD48 pKa = 3.41TAKK51 pKa = 10.53QDD53 pKa = 3.71TAEE56 pKa = 4.31PAASGDD62 pKa = 3.51AAQAADD68 pKa = 3.56ITLWTYY74 pKa = 10.79PVGSWGDD81 pKa = 3.28AATVDD86 pKa = 5.17DD87 pKa = 5.27MIANFNAAYY96 pKa = 9.4PDD98 pKa = 3.06IHH100 pKa = 6.03VTVEE104 pKa = 3.71YY105 pKa = 10.93LDD107 pKa = 3.8YY108 pKa = 11.48TNGDD112 pKa = 3.66DD113 pKa = 5.12QINTAIEE120 pKa = 4.27GNQAPDD126 pKa = 3.07IVLEE130 pKa = 4.45GPEE133 pKa = 3.97RR134 pKa = 11.84LVANWGARR142 pKa = 11.84GLMVDD147 pKa = 5.69LADD150 pKa = 5.15LWTDD154 pKa = 3.38DD155 pKa = 4.33AKK157 pKa = 11.28AAIYY161 pKa = 10.52DD162 pKa = 4.05SVEE165 pKa = 4.13NACKK169 pKa = 10.56SSDD172 pKa = 3.39GVFYY176 pKa = 9.91EE177 pKa = 4.79YY178 pKa = 9.82PLCMTAHH185 pKa = 6.13TMAINRR191 pKa = 11.84DD192 pKa = 3.06IFEE195 pKa = 4.45AAGALQYY202 pKa = 11.12LDD204 pKa = 4.29EE205 pKa = 5.21EE206 pKa = 4.66KK207 pKa = 10.62GTWTTEE213 pKa = 3.67NFQKK217 pKa = 10.68AVQAVYY223 pKa = 11.15DD224 pKa = 3.79NGQQNVGAVYY234 pKa = 10.63CSGQGGDD241 pKa = 2.96QGTRR245 pKa = 11.84ALINNLYY252 pKa = 10.63GGTFTNPEE260 pKa = 4.01HH261 pKa = 6.41TEE263 pKa = 3.87YY264 pKa = 10.23TANSPEE270 pKa = 4.19NIKK273 pKa = 10.73ALEE276 pKa = 4.21LLKK279 pKa = 11.38SMDD282 pKa = 4.81GINFDD287 pKa = 3.76ASIAGGDD294 pKa = 4.01EE295 pKa = 4.08VNMFCNGTFAMAFCWNATQEE315 pKa = 4.56KK316 pKa = 10.3NNQEE320 pKa = 3.74QGNINFDD327 pKa = 4.2VLPMAFPSEE336 pKa = 4.07NGEE339 pKa = 3.95PQLCGGIWGFGIFDD353 pKa = 4.06NGDD356 pKa = 3.41AAKK359 pKa = 10.07IAASKK364 pKa = 9.63TFIDD368 pKa = 5.9FMANDD373 pKa = 3.92PTQAPLSVQASNFWPVKK390 pKa = 10.27DD391 pKa = 4.76LGNIYY396 pKa = 10.84DD397 pKa = 4.52GDD399 pKa = 4.25DD400 pKa = 4.06LMSEE404 pKa = 4.23YY405 pKa = 11.26AKK407 pKa = 10.3FIPYY411 pKa = 9.15MGDD414 pKa = 3.18YY415 pKa = 8.96YY416 pKa = 11.11QVVGGWAEE424 pKa = 4.84ARR426 pKa = 11.84TAWWNMLQQIGSGTDD441 pKa = 2.74VATAVEE447 pKa = 4.49EE448 pKa = 4.5FVTTANAAAAKK459 pKa = 10.05
MM1 pKa = 7.84KK2 pKa = 10.01KK3 pKa = 10.32RR4 pKa = 11.84KK5 pKa = 8.19ITALLMSLVMVCSLTACGGKK25 pKa = 8.34DD26 pKa = 3.07TTTTQPEE33 pKa = 4.38PAPAPAPADD42 pKa = 3.41NAATQDD48 pKa = 3.41TAKK51 pKa = 10.53QDD53 pKa = 3.71TAEE56 pKa = 4.31PAASGDD62 pKa = 3.51AAQAADD68 pKa = 3.56ITLWTYY74 pKa = 10.79PVGSWGDD81 pKa = 3.28AATVDD86 pKa = 5.17DD87 pKa = 5.27MIANFNAAYY96 pKa = 9.4PDD98 pKa = 3.06IHH100 pKa = 6.03VTVEE104 pKa = 3.71YY105 pKa = 10.93LDD107 pKa = 3.8YY108 pKa = 11.48TNGDD112 pKa = 3.66DD113 pKa = 5.12QINTAIEE120 pKa = 4.27GNQAPDD126 pKa = 3.07IVLEE130 pKa = 4.45GPEE133 pKa = 3.97RR134 pKa = 11.84LVANWGARR142 pKa = 11.84GLMVDD147 pKa = 5.69LADD150 pKa = 5.15LWTDD154 pKa = 3.38DD155 pKa = 4.33AKK157 pKa = 11.28AAIYY161 pKa = 10.52DD162 pKa = 4.05SVEE165 pKa = 4.13NACKK169 pKa = 10.56SSDD172 pKa = 3.39GVFYY176 pKa = 9.91EE177 pKa = 4.79YY178 pKa = 9.82PLCMTAHH185 pKa = 6.13TMAINRR191 pKa = 11.84DD192 pKa = 3.06IFEE195 pKa = 4.45AAGALQYY202 pKa = 11.12LDD204 pKa = 4.29EE205 pKa = 5.21EE206 pKa = 4.66KK207 pKa = 10.62GTWTTEE213 pKa = 3.67NFQKK217 pKa = 10.68AVQAVYY223 pKa = 11.15DD224 pKa = 3.79NGQQNVGAVYY234 pKa = 10.63CSGQGGDD241 pKa = 2.96QGTRR245 pKa = 11.84ALINNLYY252 pKa = 10.63GGTFTNPEE260 pKa = 4.01HH261 pKa = 6.41TEE263 pKa = 3.87YY264 pKa = 10.23TANSPEE270 pKa = 4.19NIKK273 pKa = 10.73ALEE276 pKa = 4.21LLKK279 pKa = 11.38SMDD282 pKa = 4.81GINFDD287 pKa = 3.76ASIAGGDD294 pKa = 4.01EE295 pKa = 4.08VNMFCNGTFAMAFCWNATQEE315 pKa = 4.56KK316 pKa = 10.3NNQEE320 pKa = 3.74QGNINFDD327 pKa = 4.2VLPMAFPSEE336 pKa = 4.07NGEE339 pKa = 3.95PQLCGGIWGFGIFDD353 pKa = 4.06NGDD356 pKa = 3.41AAKK359 pKa = 10.07IAASKK364 pKa = 9.63TFIDD368 pKa = 5.9FMANDD373 pKa = 3.92PTQAPLSVQASNFWPVKK390 pKa = 10.27DD391 pKa = 4.76LGNIYY396 pKa = 10.84DD397 pKa = 4.52GDD399 pKa = 4.25DD400 pKa = 4.06LMSEE404 pKa = 4.23YY405 pKa = 11.26AKK407 pKa = 10.3FIPYY411 pKa = 9.15MGDD414 pKa = 3.18YY415 pKa = 8.96YY416 pKa = 11.11QVVGGWAEE424 pKa = 4.84ARR426 pKa = 11.84TAWWNMLQQIGSGTDD441 pKa = 2.74VATAVEE447 pKa = 4.49EE448 pKa = 4.5FVTTANAAAAKK459 pKa = 10.05
Molecular weight: 49.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9KRQ4|R9KRQ4_9FIRM Uncharacterized protein OS=Lachnospiraceae bacterium MD335 OX=1235793 GN=C809_01967 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.95GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTKK25 pKa = 10.26AGRR28 pKa = 11.84KK29 pKa = 8.26VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.92RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.95GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTKK25 pKa = 10.26AGRR28 pKa = 11.84KK29 pKa = 8.26VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.92RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1393863 |
20 |
4709 |
302.1 |
34.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.719 ± 0.044 | 1.516 ± 0.015 |
5.857 ± 0.03 | 7.669 ± 0.038 |
4.08 ± 0.024 | 6.707 ± 0.03 |
1.633 ± 0.018 | 7.242 ± 0.037 |
6.989 ± 0.038 | 8.571 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.039 ± 0.023 | 4.734 ± 0.031 |
2.996 ± 0.023 | 3.416 ± 0.022 |
4.468 ± 0.037 | 5.769 ± 0.031 |
5.56 ± 0.047 | 6.687 ± 0.031 |
0.937 ± 0.012 | 4.411 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
