
Lake Sarah-associated circular virus-45
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.92
Get precalculated fractions of proteins
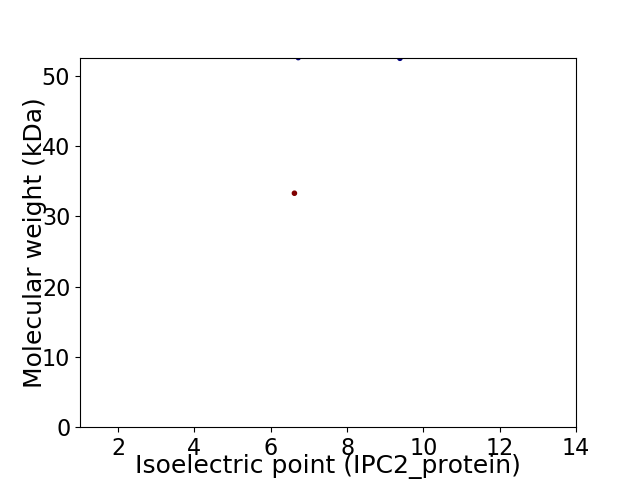
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQR0|A0A140AQR0_9VIRU Coat protein OS=Lake Sarah-associated circular virus-45 OX=1685774 PE=4 SV=1
MM1 pKa = 7.7PSFYY5 pKa = 10.48NGKK8 pKa = 9.18QFFLTYY14 pKa = 8.91PQCQHH19 pKa = 5.99SAAEE23 pKa = 4.0LAAFLTSVATCTYY36 pKa = 10.22YY37 pKa = 9.92IVAEE41 pKa = 4.35EE42 pKa = 4.04KK43 pKa = 10.89HH44 pKa = 6.34EE45 pKa = 4.86DD46 pKa = 3.8GTPHH50 pKa = 6.81LHH52 pKa = 7.11ACIQYY57 pKa = 9.68VEE59 pKa = 4.24TLRR62 pKa = 11.84GGVRR66 pKa = 11.84LLDD69 pKa = 4.1FNGHH73 pKa = 6.38HH74 pKa = 7.26PNKK77 pKa = 9.69QDD79 pKa = 3.16PRR81 pKa = 11.84KK82 pKa = 10.11FEE84 pKa = 4.01ACKK87 pKa = 10.16QYY89 pKa = 10.59CRR91 pKa = 11.84KK92 pKa = 10.21DD93 pKa = 3.28GNFIEE98 pKa = 5.38GPPEE102 pKa = 4.32AIIRR106 pKa = 11.84AAIAGLAPSDD116 pKa = 3.41IVKK119 pKa = 9.23TFTDD123 pKa = 3.27KK124 pKa = 11.36ALWFDD129 pKa = 3.13WCISKK134 pKa = 10.84RR135 pKa = 11.84ITHH138 pKa = 7.09GYY140 pKa = 9.2AEE142 pKa = 4.83WYY144 pKa = 7.2WNSTRR149 pKa = 11.84EE150 pKa = 3.78HH151 pKa = 7.36DD152 pKa = 3.79LTINEE157 pKa = 4.34DD158 pKa = 3.97TVVAGQMCEE167 pKa = 4.0SLAGYY172 pKa = 9.98AFNRR176 pKa = 11.84DD177 pKa = 3.18QHH179 pKa = 6.97RR180 pKa = 11.84VIVLKK185 pKa = 11.11GEE187 pKa = 4.29SGCGKK192 pKa = 5.88TTWAKK197 pKa = 8.25THH199 pKa = 6.23APKK202 pKa = 10.03PALFVSHH209 pKa = 7.21IDD211 pKa = 3.26SLKK214 pKa = 9.13RR215 pKa = 11.84FQPGFHH221 pKa = 6.29KK222 pKa = 9.66TIIFDD227 pKa = 4.03DD228 pKa = 3.8VDD230 pKa = 4.07FNHH233 pKa = 6.21YY234 pKa = 9.82PRR236 pKa = 11.84TGQIHH241 pKa = 5.53IVDD244 pKa = 4.19WEE246 pKa = 4.16NPRR249 pKa = 11.84AIHH252 pKa = 5.47VRR254 pKa = 11.84YY255 pKa = 8.18GTVEE259 pKa = 3.83IPAGTFKK266 pKa = 10.83IFTCNFDD273 pKa = 4.23PLTLTDD279 pKa = 3.54DD280 pKa = 5.07AIRR283 pKa = 11.84RR284 pKa = 11.84RR285 pKa = 11.84VKK287 pKa = 10.32VVNVNN292 pKa = 3.15
MM1 pKa = 7.7PSFYY5 pKa = 10.48NGKK8 pKa = 9.18QFFLTYY14 pKa = 8.91PQCQHH19 pKa = 5.99SAAEE23 pKa = 4.0LAAFLTSVATCTYY36 pKa = 10.22YY37 pKa = 9.92IVAEE41 pKa = 4.35EE42 pKa = 4.04KK43 pKa = 10.89HH44 pKa = 6.34EE45 pKa = 4.86DD46 pKa = 3.8GTPHH50 pKa = 6.81LHH52 pKa = 7.11ACIQYY57 pKa = 9.68VEE59 pKa = 4.24TLRR62 pKa = 11.84GGVRR66 pKa = 11.84LLDD69 pKa = 4.1FNGHH73 pKa = 6.38HH74 pKa = 7.26PNKK77 pKa = 9.69QDD79 pKa = 3.16PRR81 pKa = 11.84KK82 pKa = 10.11FEE84 pKa = 4.01ACKK87 pKa = 10.16QYY89 pKa = 10.59CRR91 pKa = 11.84KK92 pKa = 10.21DD93 pKa = 3.28GNFIEE98 pKa = 5.38GPPEE102 pKa = 4.32AIIRR106 pKa = 11.84AAIAGLAPSDD116 pKa = 3.41IVKK119 pKa = 9.23TFTDD123 pKa = 3.27KK124 pKa = 11.36ALWFDD129 pKa = 3.13WCISKK134 pKa = 10.84RR135 pKa = 11.84ITHH138 pKa = 7.09GYY140 pKa = 9.2AEE142 pKa = 4.83WYY144 pKa = 7.2WNSTRR149 pKa = 11.84EE150 pKa = 3.78HH151 pKa = 7.36DD152 pKa = 3.79LTINEE157 pKa = 4.34DD158 pKa = 3.97TVVAGQMCEE167 pKa = 4.0SLAGYY172 pKa = 9.98AFNRR176 pKa = 11.84DD177 pKa = 3.18QHH179 pKa = 6.97RR180 pKa = 11.84VIVLKK185 pKa = 11.11GEE187 pKa = 4.29SGCGKK192 pKa = 5.88TTWAKK197 pKa = 8.25THH199 pKa = 6.23APKK202 pKa = 10.03PALFVSHH209 pKa = 7.21IDD211 pKa = 3.26SLKK214 pKa = 9.13RR215 pKa = 11.84FQPGFHH221 pKa = 6.29KK222 pKa = 9.66TIIFDD227 pKa = 4.03DD228 pKa = 3.8VDD230 pKa = 4.07FNHH233 pKa = 6.21YY234 pKa = 9.82PRR236 pKa = 11.84TGQIHH241 pKa = 5.53IVDD244 pKa = 4.19WEE246 pKa = 4.16NPRR249 pKa = 11.84AIHH252 pKa = 5.47VRR254 pKa = 11.84YY255 pKa = 8.18GTVEE259 pKa = 3.83IPAGTFKK266 pKa = 10.83IFTCNFDD273 pKa = 4.23PLTLTDD279 pKa = 3.54DD280 pKa = 5.07AIRR283 pKa = 11.84RR284 pKa = 11.84RR285 pKa = 11.84VKK287 pKa = 10.32VVNVNN292 pKa = 3.15
Molecular weight: 33.31 kDa
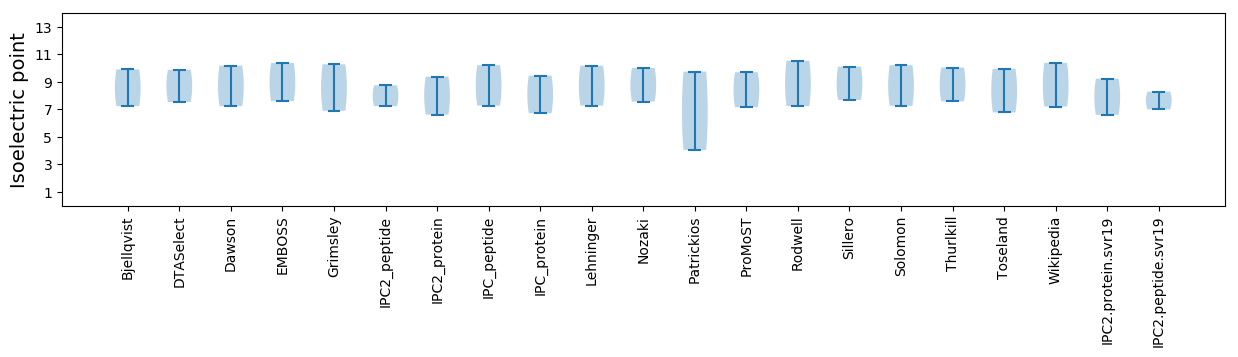
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQR0|A0A140AQR0_9VIRU Coat protein OS=Lake Sarah-associated circular virus-45 OX=1685774 PE=4 SV=1
MM1 pKa = 7.16KK2 pKa = 10.22RR3 pKa = 11.84SRR5 pKa = 11.84KK6 pKa = 8.79VAYY9 pKa = 7.55NTPPNFDD16 pKa = 3.78SPMSTSAGRR25 pKa = 11.84IISLPRR31 pKa = 11.84RR32 pKa = 11.84GRR34 pKa = 11.84GRR36 pKa = 11.84SLSGRR41 pKa = 11.84GTLTPRR47 pKa = 11.84NLFGTVAKK55 pKa = 9.14TVASAHH61 pKa = 6.2PYY63 pKa = 9.53GRR65 pKa = 11.84AALNLYY71 pKa = 9.94RR72 pKa = 11.84GARR75 pKa = 11.84AISGIFKK82 pKa = 10.41SRR84 pKa = 11.84VVNTPGLKK92 pKa = 9.93RR93 pKa = 11.84RR94 pKa = 11.84TTGGPNARR102 pKa = 11.84FVGPFKK108 pKa = 10.84KK109 pKa = 9.75PKK111 pKa = 9.97KK112 pKa = 9.39LLKK115 pKa = 10.13PSMFATKK122 pKa = 10.5GFVHH126 pKa = 6.2TCEE129 pKa = 4.18INGTVSDD136 pKa = 4.82PDD138 pKa = 3.63CVYY141 pKa = 10.36IGHH144 pKa = 5.88STFSGIMTLEE154 pKa = 4.36LLCQVLLRR162 pKa = 11.84KK163 pKa = 9.98LFAKK167 pKa = 10.53AGIVLRR173 pKa = 11.84DD174 pKa = 3.23INEE177 pKa = 4.05PLRR180 pKa = 11.84GYY182 pKa = 9.28VANTSSMWRR191 pKa = 11.84IDD193 pKa = 4.18LIRR196 pKa = 11.84HH197 pKa = 4.93NKK199 pKa = 6.28EE200 pKa = 3.71TNVYY204 pKa = 10.04DD205 pKa = 3.6VAATFTTPSATNTNIYY221 pKa = 10.02QIVGDD226 pKa = 4.34RR227 pKa = 11.84RR228 pKa = 11.84ASSNGLFPGLYY239 pKa = 8.07NTLYY243 pKa = 10.71DD244 pKa = 3.82AAVGANGSDD253 pKa = 3.88NRR255 pKa = 11.84NVWQPSSLVLYY266 pKa = 10.14QEE268 pKa = 4.88EE269 pKa = 4.25EE270 pKa = 4.19NITQFFQFRR279 pKa = 11.84AEE281 pKa = 3.77IKK283 pKa = 9.91FDD285 pKa = 3.75NEE287 pKa = 4.1TVHH290 pKa = 6.67CFSKK294 pKa = 11.01SDD296 pKa = 3.5LKK298 pKa = 10.6VQNRR302 pKa = 11.84SVSASAGTTSDD313 pKa = 4.29AVDD316 pKa = 4.01ANPLMGKK323 pKa = 10.01LYY325 pKa = 9.98HH326 pKa = 6.66FKK328 pKa = 10.76GGCPRR333 pKa = 11.84SRR335 pKa = 11.84IVNAALVEE343 pKa = 4.39SVPDD347 pKa = 3.57LSGVITARR355 pKa = 11.84AASLVSAGAQAMSEE369 pKa = 4.21PPRR372 pKa = 11.84GNAWWNCSGTSGVKK386 pKa = 9.83LQPGHH391 pKa = 6.43IKK393 pKa = 10.35YY394 pKa = 10.74DD395 pKa = 3.28SVYY398 pKa = 9.19YY399 pKa = 9.79QNAMPILKK407 pKa = 9.85FLTAMNMASGPTGKK421 pKa = 10.07QIHH424 pKa = 6.46LFGKK428 pKa = 10.33SSLLALEE435 pKa = 5.13DD436 pKa = 4.0VINVNAAQNITCSYY450 pKa = 8.16EE451 pKa = 3.77CNRR454 pKa = 11.84RR455 pKa = 11.84YY456 pKa = 10.39GIYY459 pKa = 9.47LTTKK463 pKa = 9.23TVTFAQGLKK472 pKa = 10.54YY473 pKa = 10.6AITQNDD479 pKa = 3.63LTT481 pKa = 4.4
MM1 pKa = 7.16KK2 pKa = 10.22RR3 pKa = 11.84SRR5 pKa = 11.84KK6 pKa = 8.79VAYY9 pKa = 7.55NTPPNFDD16 pKa = 3.78SPMSTSAGRR25 pKa = 11.84IISLPRR31 pKa = 11.84RR32 pKa = 11.84GRR34 pKa = 11.84GRR36 pKa = 11.84SLSGRR41 pKa = 11.84GTLTPRR47 pKa = 11.84NLFGTVAKK55 pKa = 9.14TVASAHH61 pKa = 6.2PYY63 pKa = 9.53GRR65 pKa = 11.84AALNLYY71 pKa = 9.94RR72 pKa = 11.84GARR75 pKa = 11.84AISGIFKK82 pKa = 10.41SRR84 pKa = 11.84VVNTPGLKK92 pKa = 9.93RR93 pKa = 11.84RR94 pKa = 11.84TTGGPNARR102 pKa = 11.84FVGPFKK108 pKa = 10.84KK109 pKa = 9.75PKK111 pKa = 9.97KK112 pKa = 9.39LLKK115 pKa = 10.13PSMFATKK122 pKa = 10.5GFVHH126 pKa = 6.2TCEE129 pKa = 4.18INGTVSDD136 pKa = 4.82PDD138 pKa = 3.63CVYY141 pKa = 10.36IGHH144 pKa = 5.88STFSGIMTLEE154 pKa = 4.36LLCQVLLRR162 pKa = 11.84KK163 pKa = 9.98LFAKK167 pKa = 10.53AGIVLRR173 pKa = 11.84DD174 pKa = 3.23INEE177 pKa = 4.05PLRR180 pKa = 11.84GYY182 pKa = 9.28VANTSSMWRR191 pKa = 11.84IDD193 pKa = 4.18LIRR196 pKa = 11.84HH197 pKa = 4.93NKK199 pKa = 6.28EE200 pKa = 3.71TNVYY204 pKa = 10.04DD205 pKa = 3.6VAATFTTPSATNTNIYY221 pKa = 10.02QIVGDD226 pKa = 4.34RR227 pKa = 11.84RR228 pKa = 11.84ASSNGLFPGLYY239 pKa = 8.07NTLYY243 pKa = 10.71DD244 pKa = 3.82AAVGANGSDD253 pKa = 3.88NRR255 pKa = 11.84NVWQPSSLVLYY266 pKa = 10.14QEE268 pKa = 4.88EE269 pKa = 4.25EE270 pKa = 4.19NITQFFQFRR279 pKa = 11.84AEE281 pKa = 3.77IKK283 pKa = 9.91FDD285 pKa = 3.75NEE287 pKa = 4.1TVHH290 pKa = 6.67CFSKK294 pKa = 11.01SDD296 pKa = 3.5LKK298 pKa = 10.6VQNRR302 pKa = 11.84SVSASAGTTSDD313 pKa = 4.29AVDD316 pKa = 4.01ANPLMGKK323 pKa = 10.01LYY325 pKa = 9.98HH326 pKa = 6.66FKK328 pKa = 10.76GGCPRR333 pKa = 11.84SRR335 pKa = 11.84IVNAALVEE343 pKa = 4.39SVPDD347 pKa = 3.57LSGVITARR355 pKa = 11.84AASLVSAGAQAMSEE369 pKa = 4.21PPRR372 pKa = 11.84GNAWWNCSGTSGVKK386 pKa = 9.83LQPGHH391 pKa = 6.43IKK393 pKa = 10.35YY394 pKa = 10.74DD395 pKa = 3.28SVYY398 pKa = 9.19YY399 pKa = 9.79QNAMPILKK407 pKa = 9.85FLTAMNMASGPTGKK421 pKa = 10.07QIHH424 pKa = 6.46LFGKK428 pKa = 10.33SSLLALEE435 pKa = 5.13DD436 pKa = 4.0VINVNAAQNITCSYY450 pKa = 8.16EE451 pKa = 3.77CNRR454 pKa = 11.84RR455 pKa = 11.84YY456 pKa = 10.39GIYY459 pKa = 9.47LTTKK463 pKa = 9.23TVTFAQGLKK472 pKa = 10.54YY473 pKa = 10.6AITQNDD479 pKa = 3.63LTT481 pKa = 4.4
Molecular weight: 52.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
773 |
292 |
481 |
386.5 |
42.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.668 ± 0.234 | 2.199 ± 0.46 |
4.528 ± 0.852 | 3.622 ± 0.789 |
4.916 ± 0.65 | 7.374 ± 0.63 |
2.975 ± 1.126 | 5.692 ± 0.603 |
5.563 ± 0.135 | 7.115 ± 0.852 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.552 ± 0.452 | 5.692 ± 0.824 |
5.045 ± 0.048 | 3.105 ± 0.167 |
6.21 ± 0.38 | 6.598 ± 1.653 |
7.633 ± 0.051 | 6.468 ± 0.158 |
1.294 ± 0.396 | 3.752 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
