
Phycisphaerae bacterium RAS1
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Phycisphaerae; unclassified Phycisphaerae
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
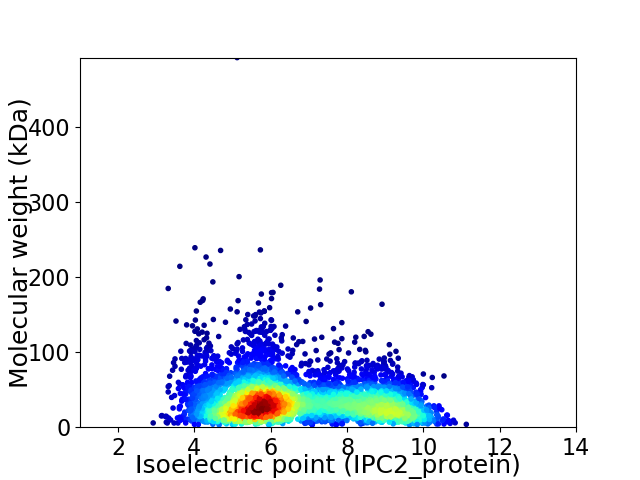
Virtual 2D-PAGE plot for 4315 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C5W7E7|A0A5C5W7E7_9BACT Co-chaperonin GroES OS=Phycisphaerae bacterium RAS1 OX=2528034 GN=RAS1_23850 PE=3 SV=1
MM1 pKa = 7.35LVCATPAAATDD12 pKa = 4.09VIDD15 pKa = 4.27CLGDD19 pKa = 3.8LVLGPAVPGFKK30 pKa = 10.14HH31 pKa = 6.47SVVADD36 pKa = 3.99PLTQATMIVGYY47 pKa = 8.03STGVTRR53 pKa = 11.84IDD55 pKa = 3.36GQYY58 pKa = 10.38GAFVICRR65 pKa = 11.84EE66 pKa = 4.13PEE68 pKa = 4.22GGVSLWVAIDD78 pKa = 3.61EE79 pKa = 4.69TTGAAALEE87 pKa = 4.3NDD89 pKa = 3.47WGADD93 pKa = 3.65LEE95 pKa = 4.65PTFQRR100 pKa = 11.84DD101 pKa = 3.27PSFGLAVADD110 pKa = 4.32WISANGAPAPGTTWDD125 pKa = 4.39LLYY128 pKa = 10.81HH129 pKa = 6.7ADD131 pKa = 3.6GVAGLGQLMGPAGAGGGDD149 pKa = 3.77PDD151 pKa = 5.49FGGVWGWLRR160 pKa = 11.84KK161 pKa = 9.68LICCLLSDD169 pKa = 4.94VPFCAPPPADD179 pKa = 5.39DD180 pKa = 5.7DD181 pKa = 5.17DD182 pKa = 7.04GDD184 pKa = 4.91GILNGDD190 pKa = 4.15DD191 pKa = 3.67NCPGVANPGQIDD203 pKa = 3.63TDD205 pKa = 3.53NDD207 pKa = 3.9GKK209 pKa = 11.63GNACDD214 pKa = 5.52DD215 pKa = 5.82DD216 pKa = 4.35IDD218 pKa = 4.93NDD220 pKa = 4.01GKK222 pKa = 10.1PNHH225 pKa = 6.29VDD227 pKa = 4.95DD228 pKa = 5.31DD229 pKa = 4.24TDD231 pKa = 5.55DD232 pKa = 5.79DD233 pKa = 5.13GDD235 pKa = 4.69PNGQDD240 pKa = 4.51DD241 pKa = 6.04DD242 pKa = 4.59IDD244 pKa = 5.78GDD246 pKa = 4.43GIPNHH251 pKa = 7.02LDD253 pKa = 2.95SDD255 pKa = 3.97MDD257 pKa = 4.16GDD259 pKa = 5.27GIPNYY264 pKa = 10.16LDD266 pKa = 3.58EE267 pKa = 7.15DD268 pKa = 3.83MDD270 pKa = 6.09GDD272 pKa = 4.2GLSNGLDD279 pKa = 3.54PDD281 pKa = 3.68IDD283 pKa = 4.22GDD285 pKa = 4.61GIPNTLDD292 pKa = 3.46PDD294 pKa = 3.63MDD296 pKa = 4.94GDD298 pKa = 4.17GATNADD304 pKa = 4.61DD305 pKa = 6.3DD306 pKa = 5.89DD307 pKa = 4.43MDD309 pKa = 6.36GDD311 pKa = 5.05GIDD314 pKa = 4.47DD315 pKa = 3.99AHH317 pKa = 7.81DD318 pKa = 3.83GDD320 pKa = 4.56IDD322 pKa = 5.61ADD324 pKa = 4.32GLPNWDD330 pKa = 5.27DD331 pKa = 3.97PDD333 pKa = 3.64MDD335 pKa = 4.32NDD337 pKa = 5.16GIANEE342 pKa = 4.96NDD344 pKa = 3.38DD345 pKa = 5.92DD346 pKa = 4.27MDD348 pKa = 6.35GDD350 pKa = 4.74GDD352 pKa = 4.94PNDD355 pKa = 5.51DD356 pKa = 5.0DD357 pKa = 6.3DD358 pKa = 6.77DD359 pKa = 5.91SEE361 pKa = 6.15GDD363 pKa = 3.45GDD365 pKa = 4.04GGGGGGALGCRR376 pKa = 11.84YY377 pKa = 9.16HH378 pKa = 7.02VCLEE382 pKa = 3.95GLRR385 pKa = 11.84LPVVSLSRR393 pKa = 11.84WPSGDD398 pKa = 3.34DD399 pKa = 3.41AQPGEE404 pKa = 5.26FGDD407 pKa = 3.65SWHH410 pKa = 6.8FGLGAEE416 pKa = 4.31PQFPCVSSIYY426 pKa = 10.21DD427 pKa = 4.02AIGLTIKK434 pKa = 10.61VAIQPPEE441 pKa = 4.16GACTRR446 pKa = 11.84DD447 pKa = 2.95AVLRR451 pKa = 11.84IRR453 pKa = 11.84ARR455 pKa = 11.84VGAAVVAEE463 pKa = 4.25GQLVFLLGAPPQDD476 pKa = 3.24RR477 pKa = 11.84ATLEE481 pKa = 4.12LATAAPLPAPAVGLLQRR498 pKa = 11.84WVVLDD503 pKa = 3.85EE504 pKa = 4.86SMDD507 pKa = 3.8GEE509 pKa = 4.11IWSPFAQATPSQWWIMVDD527 pKa = 3.42RR528 pKa = 11.84WPHH531 pKa = 4.33QLFGAIRR538 pKa = 11.84YY539 pKa = 9.4DD540 pKa = 3.61EE541 pKa = 5.36AVDD544 pKa = 4.28KK545 pKa = 11.15VAGYY549 pKa = 11.46ANGAQSYY556 pKa = 9.8SEE558 pKa = 3.65IAARR562 pKa = 11.84CTIGVDD568 pKa = 3.91SDD570 pKa = 3.2IKK572 pKa = 9.26YY573 pKa = 10.62NPMRR577 pKa = 11.84RR578 pKa = 11.84DD579 pKa = 3.32PSGHH583 pKa = 6.28ILSLYY588 pKa = 10.27RR589 pKa = 11.84LEE591 pKa = 4.62QGQCSAHH598 pKa = 6.21ARR600 pKa = 11.84LLTYY604 pKa = 10.73LLSCGGIDD612 pKa = 4.49AFTRR616 pKa = 11.84YY617 pKa = 8.65YY618 pKa = 10.32WSGSDD623 pKa = 3.11TLRR626 pKa = 11.84RR627 pKa = 11.84DD628 pKa = 3.01IYY630 pKa = 11.23NYY632 pKa = 10.31RR633 pKa = 11.84GWNATFQVEE642 pKa = 4.75RR643 pKa = 11.84PAHH646 pKa = 6.7DD647 pKa = 3.92GNEE650 pKa = 4.23LNPHH654 pKa = 6.47FSFHH658 pKa = 6.64CLTRR662 pKa = 11.84AEE664 pKa = 4.41LSGVLCDD671 pKa = 4.03PSYY674 pKa = 11.4GATGPLGILEE684 pKa = 4.67SGTAFSVDD692 pKa = 3.63AQGQPHH698 pKa = 7.06WPPAAGEE705 pKa = 4.03ALHH708 pKa = 6.38VPSILSGPAAPPEE721 pKa = 4.54RR722 pKa = 11.84IYY724 pKa = 11.5DD725 pKa = 3.59WTCPHH730 pKa = 7.02
MM1 pKa = 7.35LVCATPAAATDD12 pKa = 4.09VIDD15 pKa = 4.27CLGDD19 pKa = 3.8LVLGPAVPGFKK30 pKa = 10.14HH31 pKa = 6.47SVVADD36 pKa = 3.99PLTQATMIVGYY47 pKa = 8.03STGVTRR53 pKa = 11.84IDD55 pKa = 3.36GQYY58 pKa = 10.38GAFVICRR65 pKa = 11.84EE66 pKa = 4.13PEE68 pKa = 4.22GGVSLWVAIDD78 pKa = 3.61EE79 pKa = 4.69TTGAAALEE87 pKa = 4.3NDD89 pKa = 3.47WGADD93 pKa = 3.65LEE95 pKa = 4.65PTFQRR100 pKa = 11.84DD101 pKa = 3.27PSFGLAVADD110 pKa = 4.32WISANGAPAPGTTWDD125 pKa = 4.39LLYY128 pKa = 10.81HH129 pKa = 6.7ADD131 pKa = 3.6GVAGLGQLMGPAGAGGGDD149 pKa = 3.77PDD151 pKa = 5.49FGGVWGWLRR160 pKa = 11.84KK161 pKa = 9.68LICCLLSDD169 pKa = 4.94VPFCAPPPADD179 pKa = 5.39DD180 pKa = 5.7DD181 pKa = 5.17DD182 pKa = 7.04GDD184 pKa = 4.91GILNGDD190 pKa = 4.15DD191 pKa = 3.67NCPGVANPGQIDD203 pKa = 3.63TDD205 pKa = 3.53NDD207 pKa = 3.9GKK209 pKa = 11.63GNACDD214 pKa = 5.52DD215 pKa = 5.82DD216 pKa = 4.35IDD218 pKa = 4.93NDD220 pKa = 4.01GKK222 pKa = 10.1PNHH225 pKa = 6.29VDD227 pKa = 4.95DD228 pKa = 5.31DD229 pKa = 4.24TDD231 pKa = 5.55DD232 pKa = 5.79DD233 pKa = 5.13GDD235 pKa = 4.69PNGQDD240 pKa = 4.51DD241 pKa = 6.04DD242 pKa = 4.59IDD244 pKa = 5.78GDD246 pKa = 4.43GIPNHH251 pKa = 7.02LDD253 pKa = 2.95SDD255 pKa = 3.97MDD257 pKa = 4.16GDD259 pKa = 5.27GIPNYY264 pKa = 10.16LDD266 pKa = 3.58EE267 pKa = 7.15DD268 pKa = 3.83MDD270 pKa = 6.09GDD272 pKa = 4.2GLSNGLDD279 pKa = 3.54PDD281 pKa = 3.68IDD283 pKa = 4.22GDD285 pKa = 4.61GIPNTLDD292 pKa = 3.46PDD294 pKa = 3.63MDD296 pKa = 4.94GDD298 pKa = 4.17GATNADD304 pKa = 4.61DD305 pKa = 6.3DD306 pKa = 5.89DD307 pKa = 4.43MDD309 pKa = 6.36GDD311 pKa = 5.05GIDD314 pKa = 4.47DD315 pKa = 3.99AHH317 pKa = 7.81DD318 pKa = 3.83GDD320 pKa = 4.56IDD322 pKa = 5.61ADD324 pKa = 4.32GLPNWDD330 pKa = 5.27DD331 pKa = 3.97PDD333 pKa = 3.64MDD335 pKa = 4.32NDD337 pKa = 5.16GIANEE342 pKa = 4.96NDD344 pKa = 3.38DD345 pKa = 5.92DD346 pKa = 4.27MDD348 pKa = 6.35GDD350 pKa = 4.74GDD352 pKa = 4.94PNDD355 pKa = 5.51DD356 pKa = 5.0DD357 pKa = 6.3DD358 pKa = 6.77DD359 pKa = 5.91SEE361 pKa = 6.15GDD363 pKa = 3.45GDD365 pKa = 4.04GGGGGGALGCRR376 pKa = 11.84YY377 pKa = 9.16HH378 pKa = 7.02VCLEE382 pKa = 3.95GLRR385 pKa = 11.84LPVVSLSRR393 pKa = 11.84WPSGDD398 pKa = 3.34DD399 pKa = 3.41AQPGEE404 pKa = 5.26FGDD407 pKa = 3.65SWHH410 pKa = 6.8FGLGAEE416 pKa = 4.31PQFPCVSSIYY426 pKa = 10.21DD427 pKa = 4.02AIGLTIKK434 pKa = 10.61VAIQPPEE441 pKa = 4.16GACTRR446 pKa = 11.84DD447 pKa = 2.95AVLRR451 pKa = 11.84IRR453 pKa = 11.84ARR455 pKa = 11.84VGAAVVAEE463 pKa = 4.25GQLVFLLGAPPQDD476 pKa = 3.24RR477 pKa = 11.84ATLEE481 pKa = 4.12LATAAPLPAPAVGLLQRR498 pKa = 11.84WVVLDD503 pKa = 3.85EE504 pKa = 4.86SMDD507 pKa = 3.8GEE509 pKa = 4.11IWSPFAQATPSQWWIMVDD527 pKa = 3.42RR528 pKa = 11.84WPHH531 pKa = 4.33QLFGAIRR538 pKa = 11.84YY539 pKa = 9.4DD540 pKa = 3.61EE541 pKa = 5.36AVDD544 pKa = 4.28KK545 pKa = 11.15VAGYY549 pKa = 11.46ANGAQSYY556 pKa = 9.8SEE558 pKa = 3.65IAARR562 pKa = 11.84CTIGVDD568 pKa = 3.91SDD570 pKa = 3.2IKK572 pKa = 9.26YY573 pKa = 10.62NPMRR577 pKa = 11.84RR578 pKa = 11.84DD579 pKa = 3.32PSGHH583 pKa = 6.28ILSLYY588 pKa = 10.27RR589 pKa = 11.84LEE591 pKa = 4.62QGQCSAHH598 pKa = 6.21ARR600 pKa = 11.84LLTYY604 pKa = 10.73LLSCGGIDD612 pKa = 4.49AFTRR616 pKa = 11.84YY617 pKa = 8.65YY618 pKa = 10.32WSGSDD623 pKa = 3.11TLRR626 pKa = 11.84RR627 pKa = 11.84DD628 pKa = 3.01IYY630 pKa = 11.23NYY632 pKa = 10.31RR633 pKa = 11.84GWNATFQVEE642 pKa = 4.75RR643 pKa = 11.84PAHH646 pKa = 6.7DD647 pKa = 3.92GNEE650 pKa = 4.23LNPHH654 pKa = 6.47FSFHH658 pKa = 6.64CLTRR662 pKa = 11.84AEE664 pKa = 4.41LSGVLCDD671 pKa = 4.03PSYY674 pKa = 11.4GATGPLGILEE684 pKa = 4.67SGTAFSVDD692 pKa = 3.63AQGQPHH698 pKa = 7.06WPPAAGEE705 pKa = 4.03ALHH708 pKa = 6.38VPSILSGPAAPPEE721 pKa = 4.54RR722 pKa = 11.84IYY724 pKa = 11.5DD725 pKa = 3.59WTCPHH730 pKa = 7.02
Molecular weight: 77.08 kDa
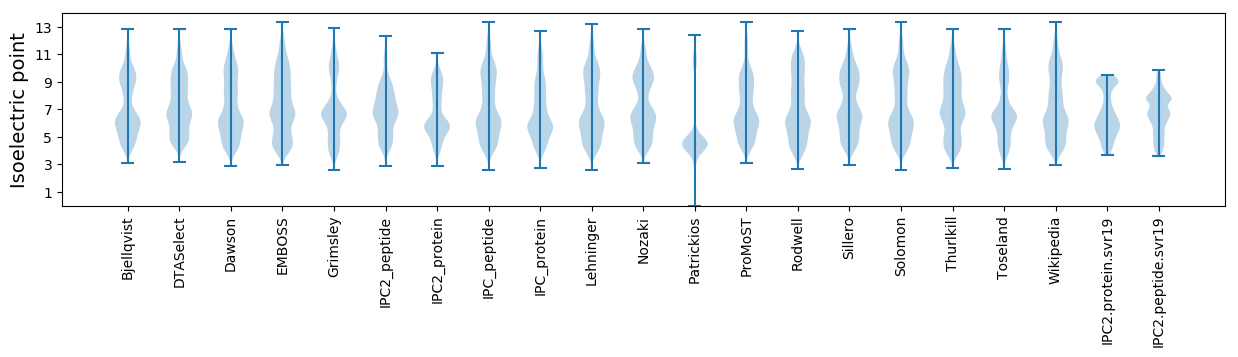
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C5W3E5|A0A5C5W3E5_9BACT Succinate dehydrogenase flavoprotein subunit OS=Phycisphaerae bacterium RAS1 OX=2528034 GN=sdhA PE=4 SV=1
MM1 pKa = 7.18NKK3 pKa = 9.67KK4 pKa = 10.07GKK6 pKa = 9.83AAAAKK11 pKa = 9.37HH12 pKa = 4.94RR13 pKa = 11.84KK14 pKa = 8.31KK15 pKa = 10.22RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84MKK20 pKa = 10.47ARR22 pKa = 11.84SKK24 pKa = 10.83ARR26 pKa = 11.84RR27 pKa = 11.84TAARR31 pKa = 11.84KK32 pKa = 9.16AA33 pKa = 3.34
MM1 pKa = 7.18NKK3 pKa = 9.67KK4 pKa = 10.07GKK6 pKa = 9.83AAAAKK11 pKa = 9.37HH12 pKa = 4.94RR13 pKa = 11.84KK14 pKa = 8.31KK15 pKa = 10.22RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84MKK20 pKa = 10.47ARR22 pKa = 11.84SKK24 pKa = 10.83ARR26 pKa = 11.84RR27 pKa = 11.84TAARR31 pKa = 11.84KK32 pKa = 9.16AA33 pKa = 3.34
Molecular weight: 3.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1565664 |
29 |
4597 |
362.8 |
39.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.271 ± 0.058 | 1.373 ± 0.021 |
5.98 ± 0.039 | 5.522 ± 0.028 |
3.503 ± 0.018 | 8.566 ± 0.04 |
1.979 ± 0.022 | 4.529 ± 0.024 |
2.785 ± 0.032 | 9.834 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.975 ± 0.016 | 2.838 ± 0.03 |
5.671 ± 0.032 | 3.27 ± 0.023 |
7.895 ± 0.045 | 5.52 ± 0.026 |
5.302 ± 0.029 | 7.366 ± 0.029 |
1.49 ± 0.018 | 2.332 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
