
Sanxia sobemo-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.7
Get precalculated fractions of proteins
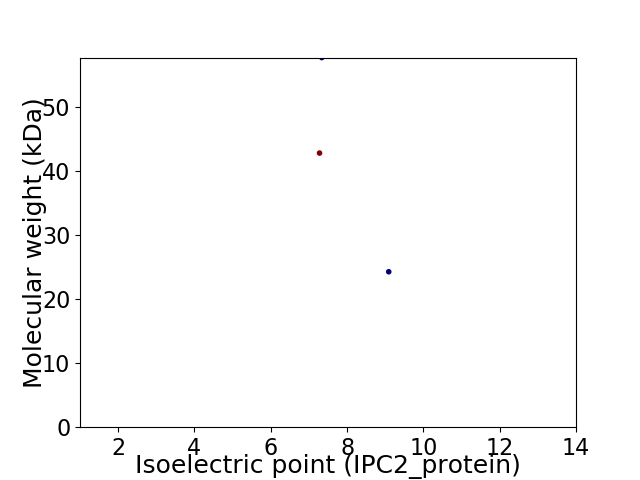
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEW3|A0A1L3KEW3_9VIRU RNA-directed RNA polymerase OS=Sanxia sobemo-like virus 1 OX=1923380 PE=4 SV=1
MM1 pKa = 7.7EE2 pKa = 4.19EE3 pKa = 4.92HH4 pKa = 6.99FDD6 pKa = 3.36QLFYY10 pKa = 11.28HH11 pKa = 6.7CVRR14 pKa = 11.84DD15 pKa = 3.95LTWTSVPGHH24 pKa = 6.71CEE26 pKa = 3.24LAKK29 pKa = 10.89YY30 pKa = 10.48GSTNAQVFDD39 pKa = 4.07LAEE42 pKa = 5.26DD43 pKa = 3.9GTLDD47 pKa = 3.34EE48 pKa = 5.7SRR50 pKa = 11.84FEE52 pKa = 4.0LVKK55 pKa = 10.57QVVYY59 pKa = 10.89LRR61 pKa = 11.84LQVLQDD67 pKa = 3.97KK68 pKa = 10.0PVADD72 pKa = 4.96PIKK75 pKa = 10.72LFIKK79 pKa = 10.54SEE81 pKa = 3.68FHH83 pKa = 6.59KK84 pKa = 10.74RR85 pKa = 11.84EE86 pKa = 4.04KK87 pKa = 10.4ILNSRR92 pKa = 11.84FRR94 pKa = 11.84LISSVSLIDD103 pKa = 3.71GLVDD107 pKa = 3.57RR108 pKa = 11.84MLFIIFAKK116 pKa = 10.52KK117 pKa = 8.89LTGNFANTGICIGYY131 pKa = 8.6NPQKK135 pKa = 10.16GGHH138 pKa = 5.76RR139 pKa = 11.84FMHH142 pKa = 5.53NTFPKK147 pKa = 9.03HH148 pKa = 4.42TEE150 pKa = 3.76KK151 pKa = 11.3LLVDD155 pKa = 3.31QTAFDD160 pKa = 3.72WTYY163 pKa = 10.44KK164 pKa = 10.17PWMANVLKK172 pKa = 10.97NVINDD177 pKa = 3.82LNMDD181 pKa = 4.02PHH183 pKa = 6.1QWWTRR188 pKa = 11.84AVDD191 pKa = 3.66NRR193 pKa = 11.84FKK195 pKa = 11.37ALFDD199 pKa = 3.34KK200 pKa = 10.95PKK202 pKa = 10.46FVFSDD207 pKa = 3.98GEE209 pKa = 4.41RR210 pKa = 11.84IDD212 pKa = 3.82QPIEE216 pKa = 3.68GVMKK220 pKa = 10.19SGCYY224 pKa = 8.44LTIIANSIAVLSIHH238 pKa = 6.0YY239 pKa = 10.08LSSLRR244 pKa = 11.84SNNPTDD250 pKa = 5.23GIILVQGDD258 pKa = 4.49DD259 pKa = 3.88AVQDD263 pKa = 3.6LPLYY267 pKa = 10.23LEE269 pKa = 4.58EE270 pKa = 4.18YY271 pKa = 10.15KK272 pKa = 10.9EE273 pKa = 4.11AVKK276 pKa = 9.79TCGVLPKK283 pKa = 10.4FKK285 pKa = 9.63TSKK288 pKa = 10.31SIEE291 pKa = 4.1FAGFRR296 pKa = 11.84YY297 pKa = 10.37EE298 pKa = 4.18NDD300 pKa = 2.74IFVPEE305 pKa = 4.17YY306 pKa = 9.88RR307 pKa = 11.84QKK309 pKa = 11.01HH310 pKa = 4.66LVALKK315 pKa = 10.41HH316 pKa = 5.36LTSDD320 pKa = 3.46RR321 pKa = 11.84LDD323 pKa = 4.3AEE325 pKa = 4.43MTLNSYY331 pKa = 10.98LRR333 pKa = 11.84MYY335 pKa = 9.86TYY337 pKa = 10.7EE338 pKa = 4.19PTMLAYY344 pKa = 9.5IRR346 pKa = 11.84SLIAKK351 pKa = 9.35RR352 pKa = 11.84NLPKK356 pKa = 10.68AHH358 pKa = 7.0LSDD361 pKa = 4.99RR362 pKa = 11.84QLKK365 pKa = 8.66WFDD368 pKa = 3.11AA369 pKa = 4.21
MM1 pKa = 7.7EE2 pKa = 4.19EE3 pKa = 4.92HH4 pKa = 6.99FDD6 pKa = 3.36QLFYY10 pKa = 11.28HH11 pKa = 6.7CVRR14 pKa = 11.84DD15 pKa = 3.95LTWTSVPGHH24 pKa = 6.71CEE26 pKa = 3.24LAKK29 pKa = 10.89YY30 pKa = 10.48GSTNAQVFDD39 pKa = 4.07LAEE42 pKa = 5.26DD43 pKa = 3.9GTLDD47 pKa = 3.34EE48 pKa = 5.7SRR50 pKa = 11.84FEE52 pKa = 4.0LVKK55 pKa = 10.57QVVYY59 pKa = 10.89LRR61 pKa = 11.84LQVLQDD67 pKa = 3.97KK68 pKa = 10.0PVADD72 pKa = 4.96PIKK75 pKa = 10.72LFIKK79 pKa = 10.54SEE81 pKa = 3.68FHH83 pKa = 6.59KK84 pKa = 10.74RR85 pKa = 11.84EE86 pKa = 4.04KK87 pKa = 10.4ILNSRR92 pKa = 11.84FRR94 pKa = 11.84LISSVSLIDD103 pKa = 3.71GLVDD107 pKa = 3.57RR108 pKa = 11.84MLFIIFAKK116 pKa = 10.52KK117 pKa = 8.89LTGNFANTGICIGYY131 pKa = 8.6NPQKK135 pKa = 10.16GGHH138 pKa = 5.76RR139 pKa = 11.84FMHH142 pKa = 5.53NTFPKK147 pKa = 9.03HH148 pKa = 4.42TEE150 pKa = 3.76KK151 pKa = 11.3LLVDD155 pKa = 3.31QTAFDD160 pKa = 3.72WTYY163 pKa = 10.44KK164 pKa = 10.17PWMANVLKK172 pKa = 10.97NVINDD177 pKa = 3.82LNMDD181 pKa = 4.02PHH183 pKa = 6.1QWWTRR188 pKa = 11.84AVDD191 pKa = 3.66NRR193 pKa = 11.84FKK195 pKa = 11.37ALFDD199 pKa = 3.34KK200 pKa = 10.95PKK202 pKa = 10.46FVFSDD207 pKa = 3.98GEE209 pKa = 4.41RR210 pKa = 11.84IDD212 pKa = 3.82QPIEE216 pKa = 3.68GVMKK220 pKa = 10.19SGCYY224 pKa = 8.44LTIIANSIAVLSIHH238 pKa = 6.0YY239 pKa = 10.08LSSLRR244 pKa = 11.84SNNPTDD250 pKa = 5.23GIILVQGDD258 pKa = 4.49DD259 pKa = 3.88AVQDD263 pKa = 3.6LPLYY267 pKa = 10.23LEE269 pKa = 4.58EE270 pKa = 4.18YY271 pKa = 10.15KK272 pKa = 10.9EE273 pKa = 4.11AVKK276 pKa = 9.79TCGVLPKK283 pKa = 10.4FKK285 pKa = 9.63TSKK288 pKa = 10.31SIEE291 pKa = 4.1FAGFRR296 pKa = 11.84YY297 pKa = 10.37EE298 pKa = 4.18NDD300 pKa = 2.74IFVPEE305 pKa = 4.17YY306 pKa = 9.88RR307 pKa = 11.84QKK309 pKa = 11.01HH310 pKa = 4.66LVALKK315 pKa = 10.41HH316 pKa = 5.36LTSDD320 pKa = 3.46RR321 pKa = 11.84LDD323 pKa = 4.3AEE325 pKa = 4.43MTLNSYY331 pKa = 10.98LRR333 pKa = 11.84MYY335 pKa = 9.86TYY337 pKa = 10.7EE338 pKa = 4.19PTMLAYY344 pKa = 9.5IRR346 pKa = 11.84SLIAKK351 pKa = 9.35RR352 pKa = 11.84NLPKK356 pKa = 10.68AHH358 pKa = 7.0LSDD361 pKa = 4.99RR362 pKa = 11.84QLKK365 pKa = 8.66WFDD368 pKa = 3.11AA369 pKa = 4.21
Molecular weight: 42.79 kDa
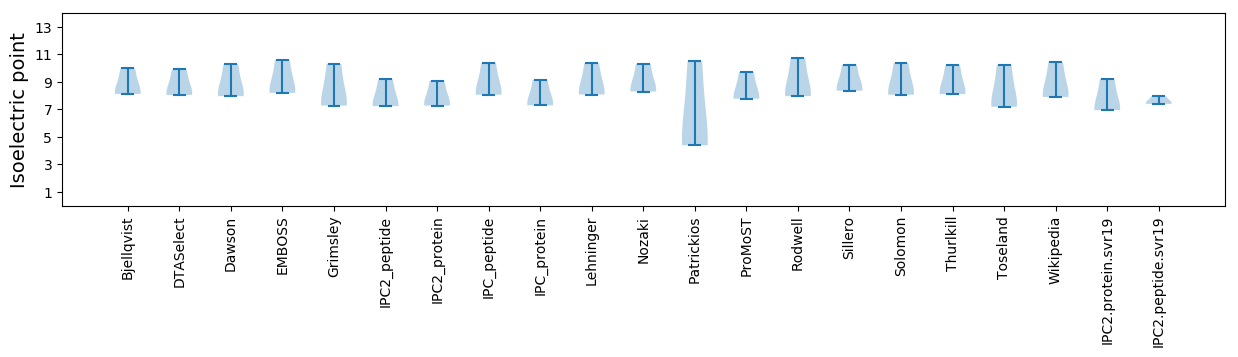
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEW3|A0A1L3KEW3_9VIRU RNA-directed RNA polymerase OS=Sanxia sobemo-like virus 1 OX=1923380 PE=4 SV=1
MM1 pKa = 6.07TTKK4 pKa = 10.23VSRR7 pKa = 11.84PDD9 pKa = 3.37QSSSAQNLLNVKK21 pKa = 10.14LEE23 pKa = 4.35SLSPTDD29 pKa = 4.03PPVLQDD35 pKa = 3.54SVIAQTFSSRR45 pKa = 11.84LNMLLMSPVNTILLAVPDD63 pKa = 4.24TDD65 pKa = 3.38TFLIQMISHH74 pKa = 6.28VFSLAILSALFYY86 pKa = 10.16PPTFGALSVWLTASRR101 pKa = 11.84SRR103 pKa = 11.84KK104 pKa = 9.83LMTVCLLLFRR114 pKa = 11.84ALKK117 pKa = 10.14RR118 pKa = 11.84HH119 pKa = 4.59QQVLCRR125 pKa = 11.84LLTTSFTMNTQARR138 pKa = 11.84PFQDD142 pKa = 3.08FLVPRR147 pKa = 11.84TCKK150 pKa = 10.36QIALRR155 pKa = 11.84EE156 pKa = 4.41CILALGPLILAFQSRR171 pKa = 11.84LLICCLIIPIWILMTWIISKK191 pKa = 10.24EE192 pKa = 3.89KK193 pKa = 10.48AISEE197 pKa = 4.06DD198 pKa = 2.93RR199 pKa = 11.84RR200 pKa = 11.84KK201 pKa = 10.39QIEE204 pKa = 4.38RR205 pKa = 11.84LLMIRR210 pKa = 11.84TLTTT214 pKa = 3.13
MM1 pKa = 6.07TTKK4 pKa = 10.23VSRR7 pKa = 11.84PDD9 pKa = 3.37QSSSAQNLLNVKK21 pKa = 10.14LEE23 pKa = 4.35SLSPTDD29 pKa = 4.03PPVLQDD35 pKa = 3.54SVIAQTFSSRR45 pKa = 11.84LNMLLMSPVNTILLAVPDD63 pKa = 4.24TDD65 pKa = 3.38TFLIQMISHH74 pKa = 6.28VFSLAILSALFYY86 pKa = 10.16PPTFGALSVWLTASRR101 pKa = 11.84SRR103 pKa = 11.84KK104 pKa = 9.83LMTVCLLLFRR114 pKa = 11.84ALKK117 pKa = 10.14RR118 pKa = 11.84HH119 pKa = 4.59QQVLCRR125 pKa = 11.84LLTTSFTMNTQARR138 pKa = 11.84PFQDD142 pKa = 3.08FLVPRR147 pKa = 11.84TCKK150 pKa = 10.36QIALRR155 pKa = 11.84EE156 pKa = 4.41CILALGPLILAFQSRR171 pKa = 11.84LLICCLIIPIWILMTWIISKK191 pKa = 10.24EE192 pKa = 3.89KK193 pKa = 10.48AISEE197 pKa = 4.06DD198 pKa = 2.93RR199 pKa = 11.84RR200 pKa = 11.84KK201 pKa = 10.39QIEE204 pKa = 4.38RR205 pKa = 11.84LLMIRR210 pKa = 11.84TLTTT214 pKa = 3.13
Molecular weight: 24.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1104 |
214 |
521 |
368.0 |
41.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.522 ± 0.467 | 1.359 ± 0.4 |
5.978 ± 0.652 | 4.348 ± 0.485 |
4.438 ± 0.751 | 4.348 ± 0.951 |
2.174 ± 0.44 | 5.616 ± 0.872 |
5.888 ± 0.706 | 11.594 ± 1.477 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.08 ± 0.258 | 3.533 ± 0.437 |
4.891 ± 0.332 | 4.438 ± 0.381 |
5.797 ± 0.315 | 8.605 ± 1.274 |
7.246 ± 0.844 | 5.978 ± 0.199 |
1.268 ± 0.19 | 2.899 ± 0.61 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
