
Human polyomavirus 7
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Deltapolyomavirus
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
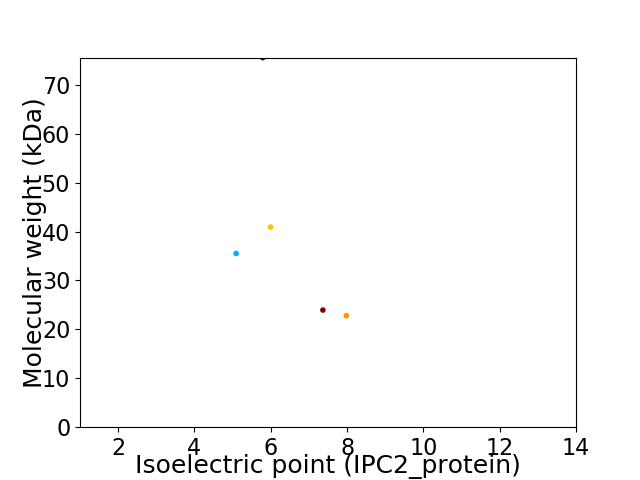
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D6QWK4|D6QWK4_9POLY VP3 OS=Human polyomavirus 7 OX=746831 PE=4 SV=1
MM1 pKa = 7.79GILLSIPEE9 pKa = 4.83IIAATAIGGGEE20 pKa = 3.99ALEE23 pKa = 4.34IAGSVGALVSGEE35 pKa = 3.95GLATLEE41 pKa = 4.35ALQSAAALSTEE52 pKa = 3.89ATAALAVSNEE62 pKa = 3.62AAIVLSTIPEE72 pKa = 4.44LSQTLFGVQTLLSSVAGVGGVVYY95 pKa = 10.58NLNPGEE101 pKa = 4.73LYY103 pKa = 10.01QAPEE107 pKa = 4.58GPGGLGPRR115 pKa = 11.84IGSTTMALQLWLPQAWPWGGAAGGVPDD142 pKa = 4.21WLLEE146 pKa = 3.97VLRR149 pKa = 11.84EE150 pKa = 4.07VPTPSEE156 pKa = 3.52ILYY159 pKa = 10.62NIARR163 pKa = 11.84GIWTSYY169 pKa = 9.21YY170 pKa = 8.51RR171 pKa = 11.84TGRR174 pKa = 11.84EE175 pKa = 3.97LIQRR179 pKa = 11.84TATRR183 pKa = 11.84EE184 pKa = 3.93LASLLSRR191 pKa = 11.84LRR193 pKa = 11.84QNIINGANRR202 pKa = 11.84AIEE205 pKa = 4.09MAPDD209 pKa = 4.13PVQGLLNLINQAIAYY224 pKa = 8.64NRR226 pKa = 11.84DD227 pKa = 2.72WEE229 pKa = 4.43TRR231 pKa = 11.84ALLEE235 pKa = 4.09GRR237 pKa = 11.84PLFEE241 pKa = 5.23PGGVVMYY248 pKa = 7.47DD249 pKa = 3.52TQNLPLSGNNDD260 pKa = 3.2QRR262 pKa = 11.84GGFHH266 pKa = 7.89DD267 pKa = 3.98EE268 pKa = 4.24GTWVSFQGEE277 pKa = 4.36EE278 pKa = 4.61GNTPQYY284 pKa = 10.0TIPQWMLFVLEE295 pKa = 4.25EE296 pKa = 4.03LQKK299 pKa = 10.89EE300 pKa = 4.4VNKK303 pKa = 10.19EE304 pKa = 3.78NKK306 pKa = 9.31HH307 pKa = 5.51ALSKK311 pKa = 9.56KK312 pKa = 10.02RR313 pKa = 11.84KK314 pKa = 4.89WTSTEE319 pKa = 3.71TSQSDD324 pKa = 3.26KK325 pKa = 10.72KK326 pKa = 10.68RR327 pKa = 11.84RR328 pKa = 11.84RR329 pKa = 3.43
MM1 pKa = 7.79GILLSIPEE9 pKa = 4.83IIAATAIGGGEE20 pKa = 3.99ALEE23 pKa = 4.34IAGSVGALVSGEE35 pKa = 3.95GLATLEE41 pKa = 4.35ALQSAAALSTEE52 pKa = 3.89ATAALAVSNEE62 pKa = 3.62AAIVLSTIPEE72 pKa = 4.44LSQTLFGVQTLLSSVAGVGGVVYY95 pKa = 10.58NLNPGEE101 pKa = 4.73LYY103 pKa = 10.01QAPEE107 pKa = 4.58GPGGLGPRR115 pKa = 11.84IGSTTMALQLWLPQAWPWGGAAGGVPDD142 pKa = 4.21WLLEE146 pKa = 3.97VLRR149 pKa = 11.84EE150 pKa = 4.07VPTPSEE156 pKa = 3.52ILYY159 pKa = 10.62NIARR163 pKa = 11.84GIWTSYY169 pKa = 9.21YY170 pKa = 8.51RR171 pKa = 11.84TGRR174 pKa = 11.84EE175 pKa = 3.97LIQRR179 pKa = 11.84TATRR183 pKa = 11.84EE184 pKa = 3.93LASLLSRR191 pKa = 11.84LRR193 pKa = 11.84QNIINGANRR202 pKa = 11.84AIEE205 pKa = 4.09MAPDD209 pKa = 4.13PVQGLLNLINQAIAYY224 pKa = 8.64NRR226 pKa = 11.84DD227 pKa = 2.72WEE229 pKa = 4.43TRR231 pKa = 11.84ALLEE235 pKa = 4.09GRR237 pKa = 11.84PLFEE241 pKa = 5.23PGGVVMYY248 pKa = 7.47DD249 pKa = 3.52TQNLPLSGNNDD260 pKa = 3.2QRR262 pKa = 11.84GGFHH266 pKa = 7.89DD267 pKa = 3.98EE268 pKa = 4.24GTWVSFQGEE277 pKa = 4.36EE278 pKa = 4.61GNTPQYY284 pKa = 10.0TIPQWMLFVLEE295 pKa = 4.25EE296 pKa = 4.03LQKK299 pKa = 10.89EE300 pKa = 4.4VNKK303 pKa = 10.19EE304 pKa = 3.78NKK306 pKa = 9.31HH307 pKa = 5.51ALSKK311 pKa = 9.56KK312 pKa = 10.02RR313 pKa = 11.84KK314 pKa = 4.89WTSTEE319 pKa = 3.71TSQSDD324 pKa = 3.26KK325 pKa = 10.72KK326 pKa = 10.68RR327 pKa = 11.84RR328 pKa = 11.84RR329 pKa = 3.43
Molecular weight: 35.52 kDa
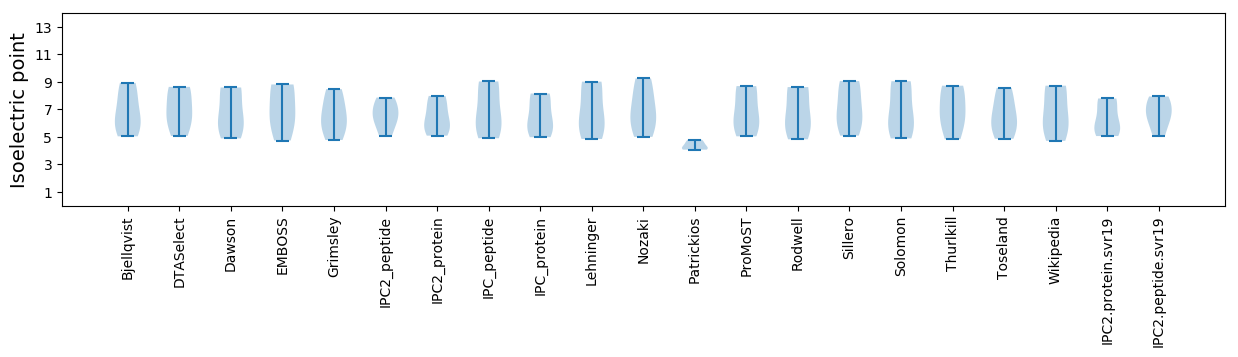
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D6QWK7|D6QWK7_9POLY Small T antigen OS=Human polyomavirus 7 OX=746831 PE=4 SV=1
MM1 pKa = 8.07DD2 pKa = 5.38KK3 pKa = 11.01LLGRR7 pKa = 11.84DD8 pKa = 3.48EE9 pKa = 4.66VKK11 pKa = 10.95EE12 pKa = 3.76LMEE15 pKa = 5.59LIGLNMACWGNLPLIQHH32 pKa = 6.46KK33 pKa = 10.38VRR35 pKa = 11.84LASKK39 pKa = 10.08KK40 pKa = 7.62YY41 pKa = 10.1HH42 pKa = 6.72PDD44 pKa = 3.38KK45 pKa = 11.5GGDD48 pKa = 3.58PQKK51 pKa = 9.93MQRR54 pKa = 11.84LNVLKK59 pKa = 10.64DD60 pKa = 3.3KK61 pKa = 11.65LEE63 pKa = 4.06ATLRR67 pKa = 11.84DD68 pKa = 3.77QRR70 pKa = 11.84SGSPMWHH77 pKa = 6.02YY78 pKa = 11.43SSDD81 pKa = 3.61EE82 pKa = 3.85VSFWDD87 pKa = 3.34IEE89 pKa = 4.34LTVGEE94 pKa = 4.22FLGPEE99 pKa = 4.39FNRR102 pKa = 11.84KK103 pKa = 8.79KK104 pKa = 9.25VWNYY108 pKa = 9.47NLCVVQGLRR117 pKa = 11.84ACCCIHH123 pKa = 7.32CILKK127 pKa = 10.42RR128 pKa = 11.84KK129 pKa = 8.92HH130 pKa = 5.68KK131 pKa = 10.48KK132 pKa = 8.78KK133 pKa = 10.31AKK135 pKa = 9.99EE136 pKa = 3.67YY137 pKa = 10.85AKK139 pKa = 10.31DD140 pKa = 3.5QRR142 pKa = 11.84GPLSWGKK149 pKa = 9.43CWCFHH154 pKa = 7.43CYY156 pKa = 10.54LNWFGVEE163 pKa = 3.77RR164 pKa = 11.84SEE166 pKa = 5.27EE167 pKa = 4.16SFMWWSHH174 pKa = 6.86IIFQTPMDD182 pKa = 4.2VLNLWGQLNLLL193 pKa = 4.33
MM1 pKa = 8.07DD2 pKa = 5.38KK3 pKa = 11.01LLGRR7 pKa = 11.84DD8 pKa = 3.48EE9 pKa = 4.66VKK11 pKa = 10.95EE12 pKa = 3.76LMEE15 pKa = 5.59LIGLNMACWGNLPLIQHH32 pKa = 6.46KK33 pKa = 10.38VRR35 pKa = 11.84LASKK39 pKa = 10.08KK40 pKa = 7.62YY41 pKa = 10.1HH42 pKa = 6.72PDD44 pKa = 3.38KK45 pKa = 11.5GGDD48 pKa = 3.58PQKK51 pKa = 9.93MQRR54 pKa = 11.84LNVLKK59 pKa = 10.64DD60 pKa = 3.3KK61 pKa = 11.65LEE63 pKa = 4.06ATLRR67 pKa = 11.84DD68 pKa = 3.77QRR70 pKa = 11.84SGSPMWHH77 pKa = 6.02YY78 pKa = 11.43SSDD81 pKa = 3.61EE82 pKa = 3.85VSFWDD87 pKa = 3.34IEE89 pKa = 4.34LTVGEE94 pKa = 4.22FLGPEE99 pKa = 4.39FNRR102 pKa = 11.84KK103 pKa = 8.79KK104 pKa = 9.25VWNYY108 pKa = 9.47NLCVVQGLRR117 pKa = 11.84ACCCIHH123 pKa = 7.32CILKK127 pKa = 10.42RR128 pKa = 11.84KK129 pKa = 8.92HH130 pKa = 5.68KK131 pKa = 10.48KK132 pKa = 8.78KK133 pKa = 10.31AKK135 pKa = 9.99EE136 pKa = 3.67YY137 pKa = 10.85AKK139 pKa = 10.31DD140 pKa = 3.5QRR142 pKa = 11.84GPLSWGKK149 pKa = 9.43CWCFHH154 pKa = 7.43CYY156 pKa = 10.54LNWFGVEE163 pKa = 3.77RR164 pKa = 11.84SEE166 pKa = 5.27EE167 pKa = 4.16SFMWWSHH174 pKa = 6.86IIFQTPMDD182 pKa = 4.2VLNLWGQLNLLL193 pKa = 4.33
Molecular weight: 22.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1781 |
193 |
671 |
356.2 |
39.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.468 ± 0.75 | 2.021 ± 0.691 |
4.323 ± 0.596 | 6.906 ± 0.505 |
3.144 ± 0.439 | 7.973 ± 0.894 |
1.46 ± 0.411 | 4.267 ± 0.315 |
6.176 ± 1.302 | 10.668 ± 0.597 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.751 ± 0.314 | 4.997 ± 0.211 |
5.783 ± 0.686 | 4.941 ± 0.537 |
4.604 ± 0.816 | 5.727 ± 0.439 |
5.334 ± 0.734 | 6.008 ± 0.561 |
2.471 ± 0.595 | 2.976 ± 0.138 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
