
Peniophora sp. CONT
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Russulales; Peniophoraceae; Peniophora; unclassified Peniophora
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
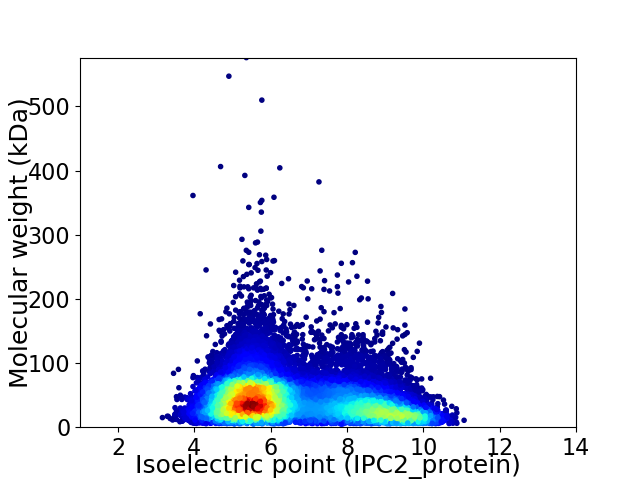
Virtual 2D-PAGE plot for 18912 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A165Z9K0|A0A165Z9K0_9AGAM Uncharacterized protein OS=Peniophora sp. CONT OX=1314672 GN=PENSPDRAFT_758779 PE=3 SV=1
MM1 pKa = 7.96ASLLPLLLLALRR13 pKa = 11.84AHH15 pKa = 7.86AYY17 pKa = 7.2TSRR20 pKa = 11.84HH21 pKa = 4.15VTFASDD27 pKa = 4.43DD28 pKa = 3.68IEE30 pKa = 4.63KK31 pKa = 9.38RR32 pKa = 11.84TALPVAQHH40 pKa = 5.69VLSLSRR46 pKa = 11.84TVPRR50 pKa = 11.84PASKK54 pKa = 10.74AKK56 pKa = 10.42ALRR59 pKa = 11.84HH60 pKa = 4.55LTNRR64 pKa = 11.84PEE66 pKa = 4.42AKK68 pKa = 9.79NSTVVSVSGSDD79 pKa = 3.17QDD81 pKa = 3.7EE82 pKa = 4.69EE83 pKa = 4.91YY84 pKa = 11.07LVDD87 pKa = 3.39ITFGTQTFPVIVDD100 pKa = 3.86TGSSDD105 pKa = 2.77TWLVAKK111 pKa = 9.75GFQCISSSGDD121 pKa = 3.44DD122 pKa = 3.9EE123 pKa = 5.14PEE125 pKa = 4.11SEE127 pKa = 4.82CDD129 pKa = 3.89FGSSGYY135 pKa = 10.37DD136 pKa = 3.17PNVSSTFVAYY146 pKa = 9.46PGHH149 pKa = 6.45NFNIAYY155 pKa = 10.11GDD157 pKa = 4.1GEE159 pKa = 4.71FLTGDD164 pKa = 3.54VGFEE168 pKa = 4.06TVTIGDD174 pKa = 4.15LSVTSQEE181 pKa = 4.15VGVVTTAFWFGDD193 pKa = 4.06SVSSGLMGLCYY204 pKa = 10.03PGLTSVFNTTDD215 pKa = 3.68PDD217 pKa = 4.31DD218 pKa = 5.47DD219 pKa = 4.37SSSNNEE225 pKa = 3.68EE226 pKa = 3.58YY227 pKa = 11.02DD228 pKa = 3.51PFFFTAIKK236 pKa = 9.75EE237 pKa = 4.49GAVSQPVFSVALNRR251 pKa = 11.84GSVAAEE257 pKa = 3.96EE258 pKa = 4.92GSVVDD263 pKa = 4.29PDD265 pKa = 4.12LGFLAFGGIAPVDD278 pKa = 3.68VTATSVTVPIQGEE291 pKa = 4.53QVSSTDD297 pKa = 3.31SEE299 pKa = 4.69FFFYY303 pKa = 10.48TVDD306 pKa = 2.72IDD308 pKa = 4.85AYY310 pKa = 10.42VFPGSDD316 pKa = 3.39ALSTSGQSILDD327 pKa = 3.5TGTTLLYY334 pKa = 10.75LPTPIANALAAAFDD348 pKa = 4.41PPAVWSDD355 pKa = 3.61DD356 pKa = 3.33EE357 pKa = 4.48GAYY360 pKa = 10.34LVNCTATAPEE370 pKa = 4.02FAVVIGGVSFNVTAADD386 pKa = 4.86LILPIGADD394 pKa = 3.14EE395 pKa = 5.91DD396 pKa = 4.49GNQVCISGHH405 pKa = 6.85DD406 pKa = 3.7DD407 pKa = 3.79GGPDD411 pKa = 3.28EE412 pKa = 5.72DD413 pKa = 4.46GNIFILGDD421 pKa = 3.68TFLHH425 pKa = 5.75NVVSTFDD432 pKa = 3.69LANNTVTLTEE442 pKa = 4.06RR443 pKa = 11.84APYY446 pKa = 9.88
MM1 pKa = 7.96ASLLPLLLLALRR13 pKa = 11.84AHH15 pKa = 7.86AYY17 pKa = 7.2TSRR20 pKa = 11.84HH21 pKa = 4.15VTFASDD27 pKa = 4.43DD28 pKa = 3.68IEE30 pKa = 4.63KK31 pKa = 9.38RR32 pKa = 11.84TALPVAQHH40 pKa = 5.69VLSLSRR46 pKa = 11.84TVPRR50 pKa = 11.84PASKK54 pKa = 10.74AKK56 pKa = 10.42ALRR59 pKa = 11.84HH60 pKa = 4.55LTNRR64 pKa = 11.84PEE66 pKa = 4.42AKK68 pKa = 9.79NSTVVSVSGSDD79 pKa = 3.17QDD81 pKa = 3.7EE82 pKa = 4.69EE83 pKa = 4.91YY84 pKa = 11.07LVDD87 pKa = 3.39ITFGTQTFPVIVDD100 pKa = 3.86TGSSDD105 pKa = 2.77TWLVAKK111 pKa = 9.75GFQCISSSGDD121 pKa = 3.44DD122 pKa = 3.9EE123 pKa = 5.14PEE125 pKa = 4.11SEE127 pKa = 4.82CDD129 pKa = 3.89FGSSGYY135 pKa = 10.37DD136 pKa = 3.17PNVSSTFVAYY146 pKa = 9.46PGHH149 pKa = 6.45NFNIAYY155 pKa = 10.11GDD157 pKa = 4.1GEE159 pKa = 4.71FLTGDD164 pKa = 3.54VGFEE168 pKa = 4.06TVTIGDD174 pKa = 4.15LSVTSQEE181 pKa = 4.15VGVVTTAFWFGDD193 pKa = 4.06SVSSGLMGLCYY204 pKa = 10.03PGLTSVFNTTDD215 pKa = 3.68PDD217 pKa = 4.31DD218 pKa = 5.47DD219 pKa = 4.37SSSNNEE225 pKa = 3.68EE226 pKa = 3.58YY227 pKa = 11.02DD228 pKa = 3.51PFFFTAIKK236 pKa = 9.75EE237 pKa = 4.49GAVSQPVFSVALNRR251 pKa = 11.84GSVAAEE257 pKa = 3.96EE258 pKa = 4.92GSVVDD263 pKa = 4.29PDD265 pKa = 4.12LGFLAFGGIAPVDD278 pKa = 3.68VTATSVTVPIQGEE291 pKa = 4.53QVSSTDD297 pKa = 3.31SEE299 pKa = 4.69FFFYY303 pKa = 10.48TVDD306 pKa = 2.72IDD308 pKa = 4.85AYY310 pKa = 10.42VFPGSDD316 pKa = 3.39ALSTSGQSILDD327 pKa = 3.5TGTTLLYY334 pKa = 10.75LPTPIANALAAAFDD348 pKa = 4.41PPAVWSDD355 pKa = 3.61DD356 pKa = 3.33EE357 pKa = 4.48GAYY360 pKa = 10.34LVNCTATAPEE370 pKa = 4.02FAVVIGGVSFNVTAADD386 pKa = 4.86LILPIGADD394 pKa = 3.14EE395 pKa = 5.91DD396 pKa = 4.49GNQVCISGHH405 pKa = 6.85DD406 pKa = 3.7DD407 pKa = 3.79GGPDD411 pKa = 3.28EE412 pKa = 5.72DD413 pKa = 4.46GNIFILGDD421 pKa = 3.68TFLHH425 pKa = 5.75NVVSTFDD432 pKa = 3.69LANNTVTLTEE442 pKa = 4.06RR443 pKa = 11.84APYY446 pKa = 9.88
Molecular weight: 46.99 kDa
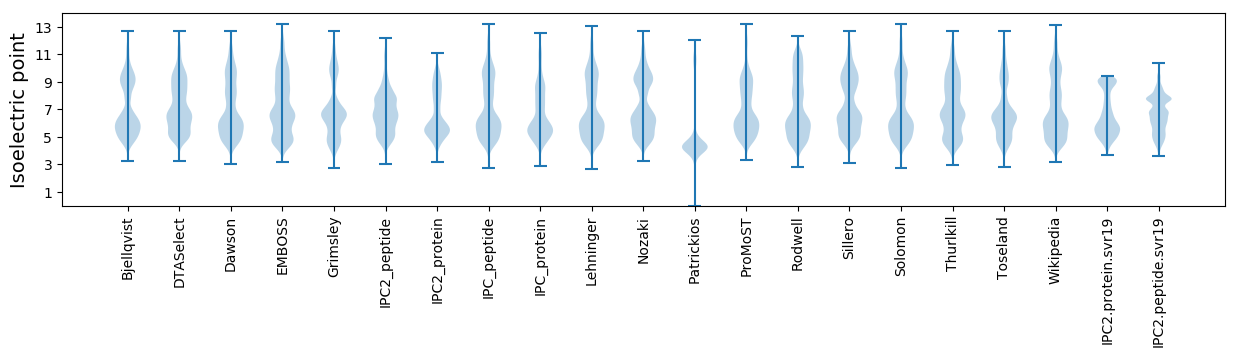
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A166I437|A0A166I437_9AGAM Uncharacterized protein OS=Peniophora sp. CONT OX=1314672 GN=PENSPDRAFT_649276 PE=3 SV=1
MM1 pKa = 7.87RR2 pKa = 11.84PTASSFNTTSNSPTSAPTYY21 pKa = 7.27TLPRR25 pKa = 11.84FFRR28 pKa = 11.84SSATAFLTRR37 pKa = 11.84TSCSRR42 pKa = 11.84RR43 pKa = 11.84VATTRR48 pKa = 11.84STTSSRR54 pKa = 11.84RR55 pKa = 11.84SATRR59 pKa = 11.84PTLLARR65 pKa = 11.84AIRR68 pKa = 11.84ALLARR73 pKa = 11.84SPRR76 pKa = 11.84VWVPSTPSGSRR87 pKa = 11.84TPSRR91 pKa = 11.84RR92 pKa = 11.84SFLVPRR98 pKa = 4.76
MM1 pKa = 7.87RR2 pKa = 11.84PTASSFNTTSNSPTSAPTYY21 pKa = 7.27TLPRR25 pKa = 11.84FFRR28 pKa = 11.84SSATAFLTRR37 pKa = 11.84TSCSRR42 pKa = 11.84RR43 pKa = 11.84VATTRR48 pKa = 11.84STTSSRR54 pKa = 11.84RR55 pKa = 11.84SATRR59 pKa = 11.84PTLLARR65 pKa = 11.84AIRR68 pKa = 11.84ALLARR73 pKa = 11.84SPRR76 pKa = 11.84VWVPSTPSGSRR87 pKa = 11.84TPSRR91 pKa = 11.84RR92 pKa = 11.84SFLVPRR98 pKa = 4.76
Molecular weight: 10.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
7847246 |
49 |
5293 |
414.9 |
45.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.765 ± 0.02 | 1.34 ± 0.007 |
5.692 ± 0.012 | 5.782 ± 0.016 |
3.582 ± 0.01 | 6.564 ± 0.015 |
2.64 ± 0.01 | 4.45 ± 0.013 |
3.813 ± 0.016 | 9.605 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.075 ± 0.006 | 3.064 ± 0.01 |
6.502 ± 0.024 | 3.438 ± 0.011 |
6.758 ± 0.018 | 8.279 ± 0.023 |
5.938 ± 0.011 | 6.545 ± 0.012 |
1.532 ± 0.006 | 2.636 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
