
Vibrio sp. UCD-FRSSP16_10
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Vibrio; unclassified Vibrio
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
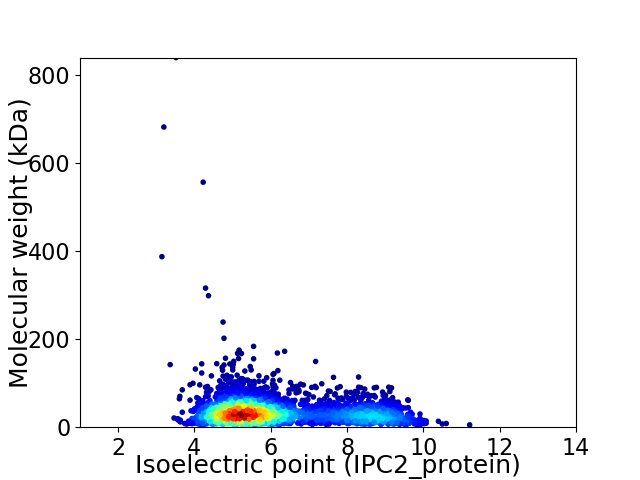
Virtual 2D-PAGE plot for 3062 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
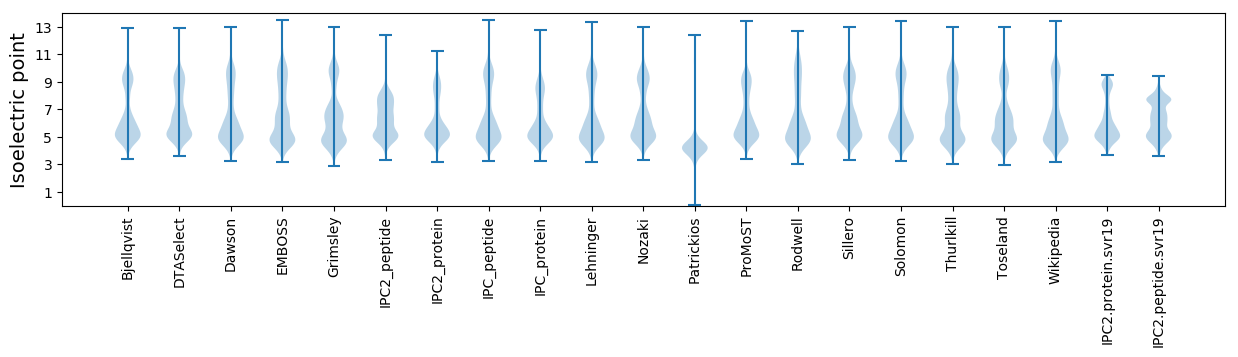
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1A6KZD8|A0A1A6KZD8_9VIBR Dehydrogenase OS=Vibrio sp. UCD-FRSSP16_10 OX=1853257 GN=A9264_02935 PE=4 SV=1
MM1 pKa = 7.17KK2 pKa = 10.07QIKK5 pKa = 9.72YY6 pKa = 10.33SAIASALLLAGISTSYY22 pKa = 10.44AAEE25 pKa = 4.91PISLTSSGDD34 pKa = 3.57DD35 pKa = 3.38GNAASNTTDD44 pKa = 3.24SDD46 pKa = 3.79LTTRR50 pKa = 11.84WSANGNDD57 pKa = 3.47GSQWVQYY64 pKa = 11.03DD65 pKa = 3.95FGSDD69 pKa = 3.35VPLSGVSIAFYY80 pKa = 10.55KK81 pKa = 10.61GDD83 pKa = 3.43SRR85 pKa = 11.84STYY88 pKa = 10.27FRR90 pKa = 11.84IEE92 pKa = 4.02SSNDD96 pKa = 2.79ASSWSTEE103 pKa = 3.59VDD105 pKa = 3.69SVSSGDD111 pKa = 3.52TADD114 pKa = 3.67QEE116 pKa = 4.89DD117 pKa = 4.25FAFSEE122 pKa = 4.94TVTARR127 pKa = 11.84YY128 pKa = 8.95IRR130 pKa = 11.84YY131 pKa = 9.51VGFGNSSNTWNSVTEE146 pKa = 3.9VDD148 pKa = 4.75FTDD151 pKa = 3.72GDD153 pKa = 4.06EE154 pKa = 4.38NVVVDD159 pKa = 5.27PDD161 pKa = 4.14PVDD164 pKa = 4.26PDD166 pKa = 3.81PVEE169 pKa = 4.52PDD171 pKa = 3.16PVVPVDD177 pKa = 3.79AAALASSDD185 pKa = 4.21DD186 pKa = 3.88GNVADD191 pKa = 5.02NVIDD195 pKa = 3.96SSLDD199 pKa = 3.59TRR201 pKa = 11.84WSANGSGEE209 pKa = 4.11WVRR212 pKa = 11.84LDD214 pKa = 3.9LGSEE218 pKa = 3.88QTLDD222 pKa = 3.31NVQLAFYY229 pKa = 9.42KK230 pKa = 10.64GSSRR234 pKa = 11.84QSIFSIEE241 pKa = 4.1VSDD244 pKa = 6.08DD245 pKa = 3.48DD246 pKa = 5.53SSWTTVLDD254 pKa = 3.54EE255 pKa = 4.78TYY257 pKa = 11.22SSGSSLALEE266 pKa = 4.17TFTFNAVNAQYY277 pKa = 10.64IRR279 pKa = 11.84YY280 pKa = 7.17TGYY283 pKa = 11.46GNTSNTWNSVTEE295 pKa = 4.21FTVSEE300 pKa = 4.31STDD303 pKa = 3.27PGGVVCEE310 pKa = 4.44EE311 pKa = 4.31GSSLVDD317 pKa = 3.64GVCVEE322 pKa = 4.85DD323 pKa = 4.19EE324 pKa = 4.56EE325 pKa = 4.79EE326 pKa = 4.21VTPITCDD333 pKa = 2.93DD334 pKa = 4.36GYY336 pKa = 11.6SLVGDD341 pKa = 3.71EE342 pKa = 4.45CVEE345 pKa = 4.31DD346 pKa = 4.05EE347 pKa = 4.78VVVIPPVTDD356 pKa = 3.56PVDD359 pKa = 3.49GQTYY363 pKa = 7.46YY364 pKa = 11.04TSADD368 pKa = 3.67DD369 pKa = 4.23LNEE372 pKa = 3.98VLASAVGGDD381 pKa = 4.13EE382 pKa = 5.71IIITDD387 pKa = 3.73SGEE390 pKa = 3.8ISIKK394 pKa = 10.56DD395 pKa = 3.27ISFDD399 pKa = 3.61SQVLIRR405 pKa = 11.84AEE407 pKa = 4.2TVGGLKK413 pKa = 10.5LEE415 pKa = 4.43NATIQNSNNITLQGFMFGPSNDD437 pKa = 2.72VSTLVKK443 pKa = 10.14IVNSTNIKK451 pKa = 9.05VLRR454 pKa = 11.84NYY456 pKa = 10.14FDD458 pKa = 4.22HH459 pKa = 7.46LDD461 pKa = 3.57VTEE464 pKa = 4.6GQSSLVVTEE473 pKa = 4.25SSQFIEE479 pKa = 3.32IGYY482 pKa = 10.02NEE484 pKa = 4.13FHH486 pKa = 7.59DD487 pKa = 4.52KK488 pKa = 10.82NISVVDD494 pKa = 3.88GAKK497 pKa = 8.89NTGSYY502 pKa = 10.5IKK504 pKa = 10.24FQYY507 pKa = 10.8DD508 pKa = 2.93VDD510 pKa = 4.02DD511 pKa = 3.97TTGEE515 pKa = 3.96EE516 pKa = 4.97LMTKK520 pKa = 9.93DD521 pKa = 3.34AHH523 pKa = 5.92IHH525 pKa = 5.91HH526 pKa = 7.1NYY528 pKa = 9.37FNNITPYY535 pKa = 10.9LVDD538 pKa = 3.82GTPAGDD544 pKa = 3.67SDD546 pKa = 4.21RR547 pKa = 11.84EE548 pKa = 4.31AIVMGISSSQDD559 pKa = 2.5IEE561 pKa = 4.33TNHH564 pKa = 5.6IVEE567 pKa = 4.36YY568 pKa = 11.12NLFEE572 pKa = 4.5NCDD575 pKa = 3.66GEE577 pKa = 5.24NEE579 pKa = 3.97ILTIKK584 pKa = 9.8TSEE587 pKa = 3.52NTFRR591 pKa = 11.84YY592 pKa = 8.23NTFKK596 pKa = 11.3NSMGSLSFRR605 pKa = 11.84LGYY608 pKa = 10.64NNSAYY613 pKa = 10.56GNYY616 pKa = 9.79FYY618 pKa = 10.76GTGASDD624 pKa = 3.87SVEE627 pKa = 3.97DD628 pKa = 5.33DD629 pKa = 3.39NYY631 pKa = 8.63QTGGIRR637 pKa = 11.84IYY639 pKa = 11.31GEE641 pKa = 3.82GHH643 pKa = 5.42SVYY646 pKa = 10.73DD647 pKa = 3.69NYY649 pKa = 10.73MEE651 pKa = 4.59GLSGTSWRR659 pKa = 11.84LPLLIDD665 pKa = 4.64NGDD668 pKa = 3.74TSDD671 pKa = 3.58SSNGDD676 pKa = 3.2SHH678 pKa = 6.26EE679 pKa = 4.31TPTNINVANNTIVDD693 pKa = 3.93STGGGIYY700 pKa = 9.7IGRR703 pKa = 11.84EE704 pKa = 3.5DD705 pKa = 3.46SSYY708 pKa = 11.63KK709 pKa = 10.41NSPSNITVTGNVVIGSEE726 pKa = 4.2GTLFGNDD733 pKa = 4.04ADD735 pKa = 4.85DD736 pKa = 5.77SSNTWSGNTAYY747 pKa = 10.66NSGSATVNSGGILDD761 pKa = 3.79TSEE764 pKa = 4.67LVEE767 pKa = 4.31LTSSPSVSKK776 pKa = 8.96PTMLTEE782 pKa = 4.05SDD784 pKa = 3.36VGINANN790 pKa = 3.57
MM1 pKa = 7.17KK2 pKa = 10.07QIKK5 pKa = 9.72YY6 pKa = 10.33SAIASALLLAGISTSYY22 pKa = 10.44AAEE25 pKa = 4.91PISLTSSGDD34 pKa = 3.57DD35 pKa = 3.38GNAASNTTDD44 pKa = 3.24SDD46 pKa = 3.79LTTRR50 pKa = 11.84WSANGNDD57 pKa = 3.47GSQWVQYY64 pKa = 11.03DD65 pKa = 3.95FGSDD69 pKa = 3.35VPLSGVSIAFYY80 pKa = 10.55KK81 pKa = 10.61GDD83 pKa = 3.43SRR85 pKa = 11.84STYY88 pKa = 10.27FRR90 pKa = 11.84IEE92 pKa = 4.02SSNDD96 pKa = 2.79ASSWSTEE103 pKa = 3.59VDD105 pKa = 3.69SVSSGDD111 pKa = 3.52TADD114 pKa = 3.67QEE116 pKa = 4.89DD117 pKa = 4.25FAFSEE122 pKa = 4.94TVTARR127 pKa = 11.84YY128 pKa = 8.95IRR130 pKa = 11.84YY131 pKa = 9.51VGFGNSSNTWNSVTEE146 pKa = 3.9VDD148 pKa = 4.75FTDD151 pKa = 3.72GDD153 pKa = 4.06EE154 pKa = 4.38NVVVDD159 pKa = 5.27PDD161 pKa = 4.14PVDD164 pKa = 4.26PDD166 pKa = 3.81PVEE169 pKa = 4.52PDD171 pKa = 3.16PVVPVDD177 pKa = 3.79AAALASSDD185 pKa = 4.21DD186 pKa = 3.88GNVADD191 pKa = 5.02NVIDD195 pKa = 3.96SSLDD199 pKa = 3.59TRR201 pKa = 11.84WSANGSGEE209 pKa = 4.11WVRR212 pKa = 11.84LDD214 pKa = 3.9LGSEE218 pKa = 3.88QTLDD222 pKa = 3.31NVQLAFYY229 pKa = 9.42KK230 pKa = 10.64GSSRR234 pKa = 11.84QSIFSIEE241 pKa = 4.1VSDD244 pKa = 6.08DD245 pKa = 3.48DD246 pKa = 5.53SSWTTVLDD254 pKa = 3.54EE255 pKa = 4.78TYY257 pKa = 11.22SSGSSLALEE266 pKa = 4.17TFTFNAVNAQYY277 pKa = 10.64IRR279 pKa = 11.84YY280 pKa = 7.17TGYY283 pKa = 11.46GNTSNTWNSVTEE295 pKa = 4.21FTVSEE300 pKa = 4.31STDD303 pKa = 3.27PGGVVCEE310 pKa = 4.44EE311 pKa = 4.31GSSLVDD317 pKa = 3.64GVCVEE322 pKa = 4.85DD323 pKa = 4.19EE324 pKa = 4.56EE325 pKa = 4.79EE326 pKa = 4.21VTPITCDD333 pKa = 2.93DD334 pKa = 4.36GYY336 pKa = 11.6SLVGDD341 pKa = 3.71EE342 pKa = 4.45CVEE345 pKa = 4.31DD346 pKa = 4.05EE347 pKa = 4.78VVVIPPVTDD356 pKa = 3.56PVDD359 pKa = 3.49GQTYY363 pKa = 7.46YY364 pKa = 11.04TSADD368 pKa = 3.67DD369 pKa = 4.23LNEE372 pKa = 3.98VLASAVGGDD381 pKa = 4.13EE382 pKa = 5.71IIITDD387 pKa = 3.73SGEE390 pKa = 3.8ISIKK394 pKa = 10.56DD395 pKa = 3.27ISFDD399 pKa = 3.61SQVLIRR405 pKa = 11.84AEE407 pKa = 4.2TVGGLKK413 pKa = 10.5LEE415 pKa = 4.43NATIQNSNNITLQGFMFGPSNDD437 pKa = 2.72VSTLVKK443 pKa = 10.14IVNSTNIKK451 pKa = 9.05VLRR454 pKa = 11.84NYY456 pKa = 10.14FDD458 pKa = 4.22HH459 pKa = 7.46LDD461 pKa = 3.57VTEE464 pKa = 4.6GQSSLVVTEE473 pKa = 4.25SSQFIEE479 pKa = 3.32IGYY482 pKa = 10.02NEE484 pKa = 4.13FHH486 pKa = 7.59DD487 pKa = 4.52KK488 pKa = 10.82NISVVDD494 pKa = 3.88GAKK497 pKa = 8.89NTGSYY502 pKa = 10.5IKK504 pKa = 10.24FQYY507 pKa = 10.8DD508 pKa = 2.93VDD510 pKa = 4.02DD511 pKa = 3.97TTGEE515 pKa = 3.96EE516 pKa = 4.97LMTKK520 pKa = 9.93DD521 pKa = 3.34AHH523 pKa = 5.92IHH525 pKa = 5.91HH526 pKa = 7.1NYY528 pKa = 9.37FNNITPYY535 pKa = 10.9LVDD538 pKa = 3.82GTPAGDD544 pKa = 3.67SDD546 pKa = 4.21RR547 pKa = 11.84EE548 pKa = 4.31AIVMGISSSQDD559 pKa = 2.5IEE561 pKa = 4.33TNHH564 pKa = 5.6IVEE567 pKa = 4.36YY568 pKa = 11.12NLFEE572 pKa = 4.5NCDD575 pKa = 3.66GEE577 pKa = 5.24NEE579 pKa = 3.97ILTIKK584 pKa = 9.8TSEE587 pKa = 3.52NTFRR591 pKa = 11.84YY592 pKa = 8.23NTFKK596 pKa = 11.3NSMGSLSFRR605 pKa = 11.84LGYY608 pKa = 10.64NNSAYY613 pKa = 10.56GNYY616 pKa = 9.79FYY618 pKa = 10.76GTGASDD624 pKa = 3.87SVEE627 pKa = 3.97DD628 pKa = 5.33DD629 pKa = 3.39NYY631 pKa = 8.63QTGGIRR637 pKa = 11.84IYY639 pKa = 11.31GEE641 pKa = 3.82GHH643 pKa = 5.42SVYY646 pKa = 10.73DD647 pKa = 3.69NYY649 pKa = 10.73MEE651 pKa = 4.59GLSGTSWRR659 pKa = 11.84LPLLIDD665 pKa = 4.64NGDD668 pKa = 3.74TSDD671 pKa = 3.58SSNGDD676 pKa = 3.2SHH678 pKa = 6.26EE679 pKa = 4.31TPTNINVANNTIVDD693 pKa = 3.93STGGGIYY700 pKa = 9.7IGRR703 pKa = 11.84EE704 pKa = 3.5DD705 pKa = 3.46SSYY708 pKa = 11.63KK709 pKa = 10.41NSPSNITVTGNVVIGSEE726 pKa = 4.2GTLFGNDD733 pKa = 4.04ADD735 pKa = 4.85DD736 pKa = 5.77SSNTWSGNTAYY747 pKa = 10.66NSGSATVNSGGILDD761 pKa = 3.79TSEE764 pKa = 4.67LVEE767 pKa = 4.31LTSSPSVSKK776 pKa = 8.96PTMLTEE782 pKa = 4.05SDD784 pKa = 3.36VGINANN790 pKa = 3.57
Molecular weight: 84.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1A6L279|A0A1A6L279_9VIBR Class II glutamine amidotransferase OS=Vibrio sp. UCD-FRSSP16_10 OX=1853257 GN=A9264_10430 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPSVLKK12 pKa = 10.49RR13 pKa = 11.84KK14 pKa = 7.97RR15 pKa = 11.84THH17 pKa = 5.89GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.44NGRR29 pKa = 11.84KK30 pKa = 9.39VINARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.22GRR40 pKa = 11.84ARR42 pKa = 11.84LSKK45 pKa = 10.83
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPSVLKK12 pKa = 10.49RR13 pKa = 11.84KK14 pKa = 7.97RR15 pKa = 11.84THH17 pKa = 5.89GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.44NGRR29 pKa = 11.84KK30 pKa = 9.39VINARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.22GRR40 pKa = 11.84ARR42 pKa = 11.84LSKK45 pKa = 10.83
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1010657 |
36 |
8186 |
330.1 |
36.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.338 ± 0.055 | 1.011 ± 0.018 |
5.746 ± 0.079 | 6.018 ± 0.057 |
4.118 ± 0.033 | 6.833 ± 0.053 |
2.266 ± 0.034 | 6.651 ± 0.037 |
5.444 ± 0.056 | 10.241 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.568 ± 0.029 | 4.373 ± 0.039 |
3.756 ± 0.035 | 4.634 ± 0.043 |
4.266 ± 0.052 | 6.898 ± 0.053 |
5.589 ± 0.066 | 7.06 ± 0.045 |
1.183 ± 0.019 | 3.005 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
