
Wabat virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
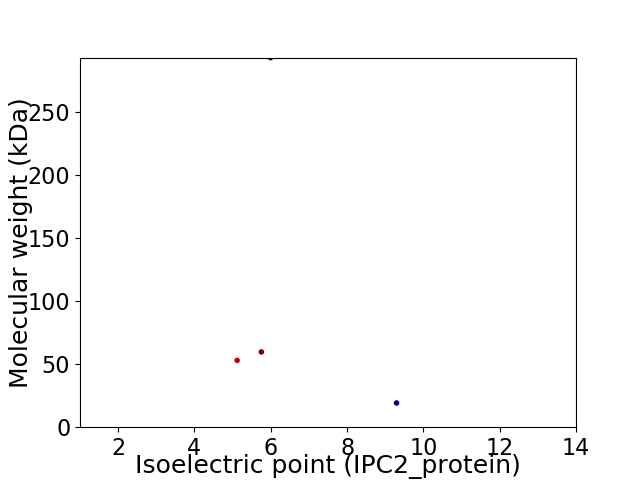
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B2RVN1|A0A1B2RVN1_9VIRU ORF4 OS=Wabat virus OX=1888308 PE=4 SV=1
MM1 pKa = 7.32FFANNDD7 pKa = 3.53QSCMRR12 pKa = 11.84TTQRR16 pKa = 11.84RR17 pKa = 11.84QSWRR21 pKa = 11.84GDD23 pKa = 3.23SKK25 pKa = 10.8QIRR28 pKa = 11.84SNLMLRR34 pKa = 11.84GSNPSTSPKK43 pKa = 10.07VSLGAQVSTTQLKK56 pKa = 8.21PQRR59 pKa = 11.84FKK61 pKa = 9.68FQKK64 pKa = 7.67MAFYY68 pKa = 10.42IPPEE72 pKa = 3.83FDD74 pKa = 2.96YY75 pKa = 11.68DD76 pKa = 3.52EE77 pKa = 4.73AKK79 pKa = 10.43RR80 pKa = 11.84EE81 pKa = 4.03RR82 pKa = 11.84EE83 pKa = 4.06LFLCKK88 pKa = 9.49NTFIQYY94 pKa = 8.67CTMGHH99 pKa = 5.94HH100 pKa = 6.58AVYY103 pKa = 10.21AYY105 pKa = 9.56IPYY108 pKa = 10.21VADD111 pKa = 3.8RR112 pKa = 11.84VDD114 pKa = 3.91KK115 pKa = 11.02EE116 pKa = 4.69HH117 pKa = 7.56IDD119 pKa = 3.36PLRR122 pKa = 11.84NFFPEE127 pKa = 4.5MIQLIEE133 pKa = 4.31RR134 pKa = 11.84SDD136 pKa = 3.67MEE138 pKa = 4.37VVRR141 pKa = 11.84DD142 pKa = 3.88TFKK145 pKa = 11.11LKK147 pKa = 9.91QGHH150 pKa = 6.4MINILSKK157 pKa = 10.77ARR159 pKa = 11.84SCMEE163 pKa = 3.7KK164 pKa = 10.32QLADD168 pKa = 4.05NLRR171 pKa = 11.84KK172 pKa = 9.74IYY174 pKa = 10.72VYY176 pKa = 10.6LARR179 pKa = 11.84DD180 pKa = 4.11LLDD183 pKa = 3.96QLQPWFVNRR192 pKa = 11.84AMEE195 pKa = 4.5TFDD198 pKa = 4.44EE199 pKa = 4.34DD200 pKa = 3.54HH201 pKa = 6.82LRR203 pKa = 11.84PTTVPPPYY211 pKa = 9.87QEE213 pKa = 5.05GGALPSYY220 pKa = 10.29EE221 pKa = 4.82SISEE225 pKa = 4.23VPQTRR230 pKa = 11.84DD231 pKa = 2.49SCTQIEE237 pKa = 4.32AHH239 pKa = 7.42DD240 pKa = 3.84MQQWEE245 pKa = 4.7DD246 pKa = 3.61FLNPRR251 pKa = 11.84DD252 pKa = 3.98EE253 pKa = 4.57ILLGDD258 pKa = 4.23HH259 pKa = 7.12DD260 pKa = 5.96GIWAHH265 pKa = 6.34YY266 pKa = 7.01EE267 pKa = 3.95QQQGEE272 pKa = 4.43LWIDD276 pKa = 3.58ATPDD280 pKa = 4.24FYY282 pKa = 11.0PEE284 pKa = 4.06TPTRR288 pKa = 11.84AIWEE292 pKa = 4.11NFRR295 pKa = 11.84TVNNLRR301 pKa = 11.84DD302 pKa = 3.9APPEE306 pKa = 3.85YY307 pKa = 10.15RR308 pKa = 11.84DD309 pKa = 3.39VLLFILTEE317 pKa = 4.1QYY319 pKa = 10.81HH320 pKa = 6.62PEE322 pKa = 3.77LTLAQDD328 pKa = 4.06YY329 pKa = 9.55EE330 pKa = 4.42HH331 pKa = 7.4TDD333 pKa = 2.98TDD335 pKa = 4.31EE336 pKa = 3.89IRR338 pKa = 11.84SIARR342 pKa = 11.84RR343 pKa = 11.84CVMWHH348 pKa = 5.55YY349 pKa = 10.36RR350 pKa = 11.84DD351 pKa = 4.97CGFCMLGLVHH361 pKa = 5.94CHH363 pKa = 6.08CRR365 pKa = 11.84ANCNCNDD372 pKa = 4.48RR373 pKa = 11.84SCVWNWLEE381 pKa = 4.83ADD383 pKa = 3.26INSYY387 pKa = 8.4LTAPNGMFYY396 pKa = 10.52EE397 pKa = 4.23LRR399 pKa = 11.84SLVMYY404 pKa = 10.11IEE406 pKa = 4.2IMGAQEE412 pKa = 5.7QNDD415 pKa = 3.22NWDD418 pKa = 3.22RR419 pKa = 11.84WFDD422 pKa = 3.79SYY424 pKa = 11.92VGATVAGHH432 pKa = 5.39SCFWSDD438 pKa = 2.78AFEE441 pKa = 3.95WTRR444 pKa = 11.84RR445 pKa = 11.84SMSS448 pKa = 3.06
MM1 pKa = 7.32FFANNDD7 pKa = 3.53QSCMRR12 pKa = 11.84TTQRR16 pKa = 11.84RR17 pKa = 11.84QSWRR21 pKa = 11.84GDD23 pKa = 3.23SKK25 pKa = 10.8QIRR28 pKa = 11.84SNLMLRR34 pKa = 11.84GSNPSTSPKK43 pKa = 10.07VSLGAQVSTTQLKK56 pKa = 8.21PQRR59 pKa = 11.84FKK61 pKa = 9.68FQKK64 pKa = 7.67MAFYY68 pKa = 10.42IPPEE72 pKa = 3.83FDD74 pKa = 2.96YY75 pKa = 11.68DD76 pKa = 3.52EE77 pKa = 4.73AKK79 pKa = 10.43RR80 pKa = 11.84EE81 pKa = 4.03RR82 pKa = 11.84EE83 pKa = 4.06LFLCKK88 pKa = 9.49NTFIQYY94 pKa = 8.67CTMGHH99 pKa = 5.94HH100 pKa = 6.58AVYY103 pKa = 10.21AYY105 pKa = 9.56IPYY108 pKa = 10.21VADD111 pKa = 3.8RR112 pKa = 11.84VDD114 pKa = 3.91KK115 pKa = 11.02EE116 pKa = 4.69HH117 pKa = 7.56IDD119 pKa = 3.36PLRR122 pKa = 11.84NFFPEE127 pKa = 4.5MIQLIEE133 pKa = 4.31RR134 pKa = 11.84SDD136 pKa = 3.67MEE138 pKa = 4.37VVRR141 pKa = 11.84DD142 pKa = 3.88TFKK145 pKa = 11.11LKK147 pKa = 9.91QGHH150 pKa = 6.4MINILSKK157 pKa = 10.77ARR159 pKa = 11.84SCMEE163 pKa = 3.7KK164 pKa = 10.32QLADD168 pKa = 4.05NLRR171 pKa = 11.84KK172 pKa = 9.74IYY174 pKa = 10.72VYY176 pKa = 10.6LARR179 pKa = 11.84DD180 pKa = 4.11LLDD183 pKa = 3.96QLQPWFVNRR192 pKa = 11.84AMEE195 pKa = 4.5TFDD198 pKa = 4.44EE199 pKa = 4.34DD200 pKa = 3.54HH201 pKa = 6.82LRR203 pKa = 11.84PTTVPPPYY211 pKa = 9.87QEE213 pKa = 5.05GGALPSYY220 pKa = 10.29EE221 pKa = 4.82SISEE225 pKa = 4.23VPQTRR230 pKa = 11.84DD231 pKa = 2.49SCTQIEE237 pKa = 4.32AHH239 pKa = 7.42DD240 pKa = 3.84MQQWEE245 pKa = 4.7DD246 pKa = 3.61FLNPRR251 pKa = 11.84DD252 pKa = 3.98EE253 pKa = 4.57ILLGDD258 pKa = 4.23HH259 pKa = 7.12DD260 pKa = 5.96GIWAHH265 pKa = 6.34YY266 pKa = 7.01EE267 pKa = 3.95QQQGEE272 pKa = 4.43LWIDD276 pKa = 3.58ATPDD280 pKa = 4.24FYY282 pKa = 11.0PEE284 pKa = 4.06TPTRR288 pKa = 11.84AIWEE292 pKa = 4.11NFRR295 pKa = 11.84TVNNLRR301 pKa = 11.84DD302 pKa = 3.9APPEE306 pKa = 3.85YY307 pKa = 10.15RR308 pKa = 11.84DD309 pKa = 3.39VLLFILTEE317 pKa = 4.1QYY319 pKa = 10.81HH320 pKa = 6.62PEE322 pKa = 3.77LTLAQDD328 pKa = 4.06YY329 pKa = 9.55EE330 pKa = 4.42HH331 pKa = 7.4TDD333 pKa = 2.98TDD335 pKa = 4.31EE336 pKa = 3.89IRR338 pKa = 11.84SIARR342 pKa = 11.84RR343 pKa = 11.84CVMWHH348 pKa = 5.55YY349 pKa = 10.36RR350 pKa = 11.84DD351 pKa = 4.97CGFCMLGLVHH361 pKa = 5.94CHH363 pKa = 6.08CRR365 pKa = 11.84ANCNCNDD372 pKa = 4.48RR373 pKa = 11.84SCVWNWLEE381 pKa = 4.83ADD383 pKa = 3.26INSYY387 pKa = 8.4LTAPNGMFYY396 pKa = 10.52EE397 pKa = 4.23LRR399 pKa = 11.84SLVMYY404 pKa = 10.11IEE406 pKa = 4.2IMGAQEE412 pKa = 5.7QNDD415 pKa = 3.22NWDD418 pKa = 3.22RR419 pKa = 11.84WFDD422 pKa = 3.79SYY424 pKa = 11.92VGATVAGHH432 pKa = 5.39SCFWSDD438 pKa = 2.78AFEE441 pKa = 3.95WTRR444 pKa = 11.84RR445 pKa = 11.84SMSS448 pKa = 3.06
Molecular weight: 52.97 kDa
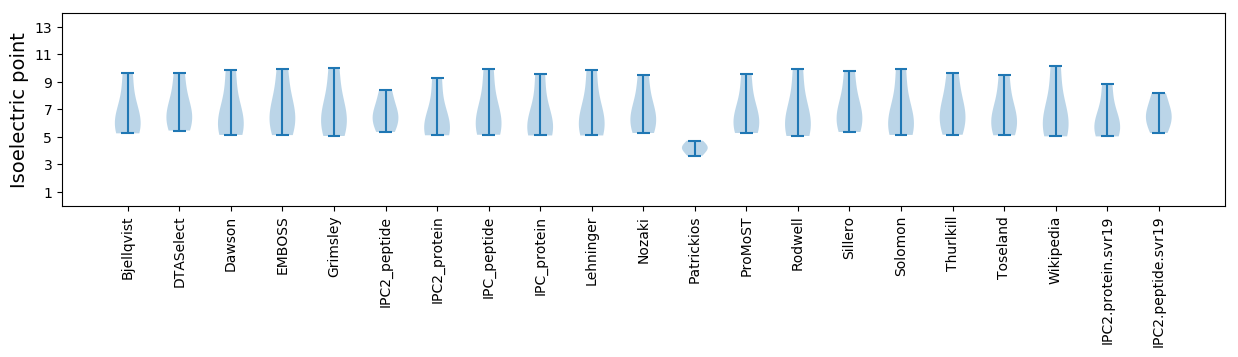
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B2RVM9|A0A1B2RVM9_9VIRU Capsid protein OS=Wabat virus OX=1888308 PE=4 SV=1
MM1 pKa = 7.03QAIAGLAGALPNTLGMGLDD20 pKa = 3.59TMNRR24 pKa = 11.84ALDD27 pKa = 3.84RR28 pKa = 11.84NQQNSQFRR36 pKa = 11.84ARR38 pKa = 11.84EE39 pKa = 3.8EE40 pKa = 4.22AFTKK44 pKa = 10.62AGLPAFLAYY53 pKa = 9.37TGAAGQPYY61 pKa = 9.49YY62 pKa = 9.95PSSYY66 pKa = 10.48SNVGGRR72 pKa = 11.84TNYY75 pKa = 7.81RR76 pKa = 11.84TGMVGSPYY84 pKa = 9.86TGQSNSLFTNAEE96 pKa = 3.95AVTNTSLGSSSPSGEE111 pKa = 4.19GPPQPMTTPTYY122 pKa = 10.15TSISSTRR129 pKa = 11.84SGFNRR134 pKa = 11.84MQGPRR139 pKa = 11.84FLKK142 pKa = 10.58DD143 pKa = 2.92VLRR146 pKa = 11.84EE147 pKa = 4.0QRR149 pKa = 11.84PEE151 pKa = 3.94LYY153 pKa = 10.49EE154 pKa = 4.24NNATQAEE161 pKa = 4.61LAWGLKK167 pKa = 9.83ADD169 pKa = 5.58PIQPHH174 pKa = 5.7VKK176 pKa = 10.24RR177 pKa = 11.84KK178 pKa = 9.43
MM1 pKa = 7.03QAIAGLAGALPNTLGMGLDD20 pKa = 3.59TMNRR24 pKa = 11.84ALDD27 pKa = 3.84RR28 pKa = 11.84NQQNSQFRR36 pKa = 11.84ARR38 pKa = 11.84EE39 pKa = 3.8EE40 pKa = 4.22AFTKK44 pKa = 10.62AGLPAFLAYY53 pKa = 9.37TGAAGQPYY61 pKa = 9.49YY62 pKa = 9.95PSSYY66 pKa = 10.48SNVGGRR72 pKa = 11.84TNYY75 pKa = 7.81RR76 pKa = 11.84TGMVGSPYY84 pKa = 9.86TGQSNSLFTNAEE96 pKa = 3.95AVTNTSLGSSSPSGEE111 pKa = 4.19GPPQPMTTPTYY122 pKa = 10.15TSISSTRR129 pKa = 11.84SGFNRR134 pKa = 11.84MQGPRR139 pKa = 11.84FLKK142 pKa = 10.58DD143 pKa = 2.92VLRR146 pKa = 11.84EE147 pKa = 4.0QRR149 pKa = 11.84PEE151 pKa = 3.94LYY153 pKa = 10.49EE154 pKa = 4.24NNATQAEE161 pKa = 4.61LAWGLKK167 pKa = 9.83ADD169 pKa = 5.58PIQPHH174 pKa = 5.7VKK176 pKa = 10.24RR177 pKa = 11.84KK178 pKa = 9.43
Molecular weight: 19.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3740 |
178 |
2584 |
935.0 |
106.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.497 ± 0.528 | 1.925 ± 0.584 |
6.07 ± 0.482 | 6.684 ± 0.68 |
4.519 ± 0.168 | 5.428 ± 0.7 |
3.048 ± 0.314 | 5.668 ± 0.615 |
6.364 ± 1.282 | 7.701 ± 0.658 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.021 ± 0.44 | 4.626 ± 0.392 |
5.187 ± 0.412 | 3.556 ± 0.793 |
5.16 ± 0.618 | 6.898 ± 0.716 |
6.203 ± 0.349 | 6.283 ± 0.787 |
1.551 ± 0.329 | 3.61 ± 0.393 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
