
Haloterrigena jeotgali icosahedral virus 1
Taxonomy: Viruses; Varidnaviria; Helvetiavirae; Dividoviricota; Laserviricetes; Halopanivirales; Simuloviridae; Yingchengvirus; Yingchengvirus HJIV1
Average proteome isoelectric point is 5.12
Get precalculated fractions of proteins
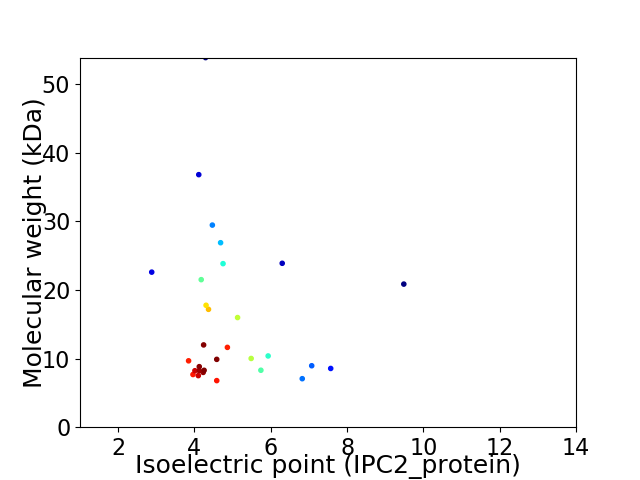
Virtual 2D-PAGE plot for 30 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7G2JX99|A0A7G2JX99_9VIRU Uncharacterized protein OS=Haloterrigena jeotgali icosahedral virus 1 OX=2766528 GN=HJIV1gp28 PE=4 SV=1
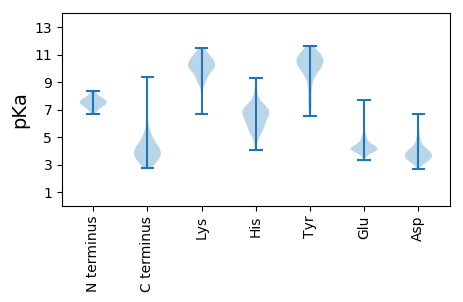
MM1 pKa = 7.08TVIGEE6 pKa = 4.13KK7 pKa = 8.97TATYY11 pKa = 7.58EE12 pKa = 4.14TLLSDD17 pKa = 5.38LVTEE21 pKa = 4.79LLTYY25 pKa = 10.64SNWSDD30 pKa = 3.19ADD32 pKa = 3.79TYY34 pKa = 9.47VTNDD38 pKa = 3.04GSDD41 pKa = 3.35TSWYY45 pKa = 10.13NNGRR49 pKa = 11.84VLQDD53 pKa = 3.38SNTGTFLLMYY63 pKa = 10.27LSGGQWHH70 pKa = 6.13EE71 pKa = 3.92QNGTQRR77 pKa = 11.84HH78 pKa = 4.06VSGIKK83 pKa = 9.92FVISTDD89 pKa = 3.03WDD91 pKa = 3.91STNHH95 pKa = 5.68YY96 pKa = 6.96PTGYY100 pKa = 8.97TNVGSQSGLEE110 pKa = 4.0ANALVSNVSNDD121 pKa = 3.2APATYY126 pKa = 10.85DD127 pKa = 4.6DD128 pKa = 5.49FNATYY133 pKa = 10.53ANHH136 pKa = 6.83SDD138 pKa = 4.32YY139 pKa = 11.02DD140 pKa = 4.2YY141 pKa = 11.61QWFASPYY148 pKa = 9.68GVWGSNNNLSRR159 pKa = 11.84DD160 pKa = 4.12TIRR163 pKa = 11.84GTSVSYY169 pKa = 10.87IMSVKK174 pKa = 10.6ADD176 pKa = 3.64GFNIGTWNSNDD187 pKa = 3.26GNNGIASVTSYY198 pKa = 10.91EE199 pKa = 4.13YY200 pKa = 11.14LSDD203 pKa = 4.85RR204 pKa = 11.84FFDD207 pKa = 4.37DD208 pKa = 4.0PGIPLIGLTRR218 pKa = 11.84TSDD221 pKa = 2.8IAQAAIYY228 pKa = 9.99GFTSYY233 pKa = 11.25SDD235 pKa = 3.37SDD237 pKa = 3.81STHH240 pKa = 5.68TNRR243 pKa = 11.84NVGFDD248 pKa = 3.55NSPVEE253 pKa = 4.17DD254 pKa = 4.64ADD256 pKa = 3.45WGIINPSSEE265 pKa = 3.98DD266 pKa = 3.08DD267 pKa = 3.63TFFFRR272 pKa = 11.84YY273 pKa = 8.19PAAFNTTAKK282 pKa = 8.68TVPVAYY288 pKa = 10.33LRR290 pKa = 11.84QTIPNDD296 pKa = 3.28SSEE299 pKa = 4.26GGAHH303 pKa = 7.14GDD305 pKa = 3.79DD306 pKa = 3.82FTHH309 pKa = 6.77NSTNYY314 pKa = 10.04RR315 pKa = 11.84VFAQSGASTSTVLSMGLRR333 pKa = 11.84HH334 pKa = 5.89EE335 pKa = 4.34
MM1 pKa = 7.08TVIGEE6 pKa = 4.13KK7 pKa = 8.97TATYY11 pKa = 7.58EE12 pKa = 4.14TLLSDD17 pKa = 5.38LVTEE21 pKa = 4.79LLTYY25 pKa = 10.64SNWSDD30 pKa = 3.19ADD32 pKa = 3.79TYY34 pKa = 9.47VTNDD38 pKa = 3.04GSDD41 pKa = 3.35TSWYY45 pKa = 10.13NNGRR49 pKa = 11.84VLQDD53 pKa = 3.38SNTGTFLLMYY63 pKa = 10.27LSGGQWHH70 pKa = 6.13EE71 pKa = 3.92QNGTQRR77 pKa = 11.84HH78 pKa = 4.06VSGIKK83 pKa = 9.92FVISTDD89 pKa = 3.03WDD91 pKa = 3.91STNHH95 pKa = 5.68YY96 pKa = 6.96PTGYY100 pKa = 8.97TNVGSQSGLEE110 pKa = 4.0ANALVSNVSNDD121 pKa = 3.2APATYY126 pKa = 10.85DD127 pKa = 4.6DD128 pKa = 5.49FNATYY133 pKa = 10.53ANHH136 pKa = 6.83SDD138 pKa = 4.32YY139 pKa = 11.02DD140 pKa = 4.2YY141 pKa = 11.61QWFASPYY148 pKa = 9.68GVWGSNNNLSRR159 pKa = 11.84DD160 pKa = 4.12TIRR163 pKa = 11.84GTSVSYY169 pKa = 10.87IMSVKK174 pKa = 10.6ADD176 pKa = 3.64GFNIGTWNSNDD187 pKa = 3.26GNNGIASVTSYY198 pKa = 10.91EE199 pKa = 4.13YY200 pKa = 11.14LSDD203 pKa = 4.85RR204 pKa = 11.84FFDD207 pKa = 4.37DD208 pKa = 4.0PGIPLIGLTRR218 pKa = 11.84TSDD221 pKa = 2.8IAQAAIYY228 pKa = 9.99GFTSYY233 pKa = 11.25SDD235 pKa = 3.37SDD237 pKa = 3.81STHH240 pKa = 5.68TNRR243 pKa = 11.84NVGFDD248 pKa = 3.55NSPVEE253 pKa = 4.17DD254 pKa = 4.64ADD256 pKa = 3.45WGIINPSSEE265 pKa = 3.98DD266 pKa = 3.08DD267 pKa = 3.63TFFFRR272 pKa = 11.84YY273 pKa = 8.19PAAFNTTAKK282 pKa = 8.68TVPVAYY288 pKa = 10.33LRR290 pKa = 11.84QTIPNDD296 pKa = 3.28SSEE299 pKa = 4.26GGAHH303 pKa = 7.14GDD305 pKa = 3.79DD306 pKa = 3.82FTHH309 pKa = 6.77NSTNYY314 pKa = 10.04RR315 pKa = 11.84VFAQSGASTSTVLSMGLRR333 pKa = 11.84HH334 pKa = 5.89EE335 pKa = 4.34
Molecular weight: 36.8 kDa
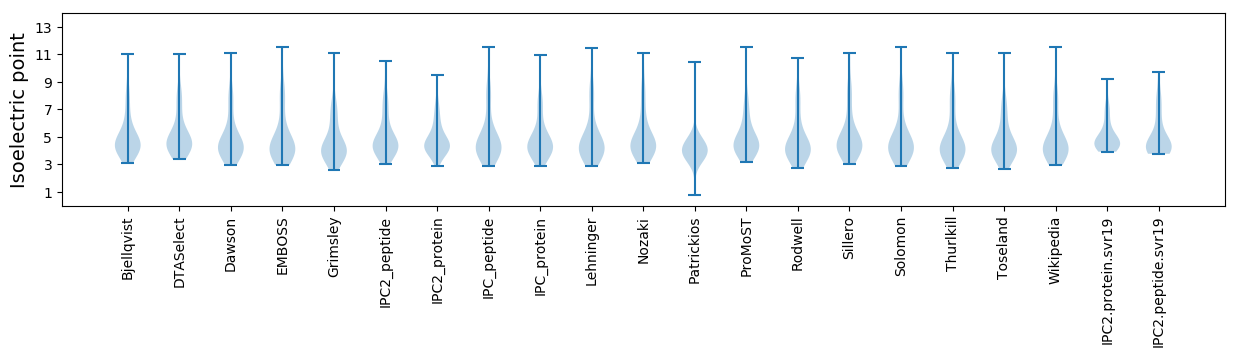
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7G2JX99|A0A7G2JX99_9VIRU Uncharacterized protein OS=Haloterrigena jeotgali icosahedral virus 1 OX=2766528 GN=HJIV1gp28 PE=4 SV=1
MM1 pKa = 7.7GLRR4 pKa = 11.84GDD6 pKa = 4.11DD7 pKa = 5.86DD8 pKa = 3.93EE9 pKa = 5.08PKK11 pKa = 10.02WRR13 pKa = 11.84LKK15 pKa = 10.05RR16 pKa = 11.84DD17 pKa = 3.36RR18 pKa = 11.84MDD20 pKa = 3.6GGEE23 pKa = 4.21LLVTRR28 pKa = 11.84RR29 pKa = 11.84DD30 pKa = 3.17GRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84AEE36 pKa = 4.11VIVGNLDD43 pKa = 3.36EE44 pKa = 4.93LVRR47 pKa = 11.84EE48 pKa = 4.23VLGNYY53 pKa = 7.33PARR56 pKa = 11.84RR57 pKa = 11.84EE58 pKa = 4.06ALEE61 pKa = 3.94AAVEE65 pKa = 4.41GWRR68 pKa = 11.84TIPTASRR75 pKa = 11.84RR76 pKa = 11.84VVLRR80 pKa = 11.84HH81 pKa = 4.25GRR83 pKa = 11.84GRR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84SSRR93 pKa = 11.84RR94 pKa = 11.84SGPAAGRR101 pKa = 11.84RR102 pKa = 11.84SSCRR106 pKa = 11.84GRR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84SSRR116 pKa = 11.84RR117 pKa = 11.84RR118 pKa = 11.84QDD120 pKa = 3.63ALLEE124 pKa = 4.28DD125 pKa = 4.93LPDD128 pKa = 4.2EE129 pKa = 5.2GDD131 pKa = 4.15DD132 pKa = 3.98GPSRR136 pKa = 11.84EE137 pKa = 4.15FRR139 pKa = 11.84AAQQDD144 pKa = 4.04GYY146 pKa = 11.15LRR148 pKa = 11.84QGDD151 pKa = 3.85SDD153 pKa = 5.0DD154 pKa = 3.88PDD156 pKa = 3.4EE157 pKa = 5.98RR158 pKa = 11.84EE159 pKa = 4.0GLGQFGARR167 pKa = 11.84RR168 pKa = 11.84SDD170 pKa = 3.44TQALEE175 pKa = 4.66DD176 pKa = 4.43FEE178 pKa = 6.97SDD180 pKa = 3.05AA181 pKa = 5.22
MM1 pKa = 7.7GLRR4 pKa = 11.84GDD6 pKa = 4.11DD7 pKa = 5.86DD8 pKa = 3.93EE9 pKa = 5.08PKK11 pKa = 10.02WRR13 pKa = 11.84LKK15 pKa = 10.05RR16 pKa = 11.84DD17 pKa = 3.36RR18 pKa = 11.84MDD20 pKa = 3.6GGEE23 pKa = 4.21LLVTRR28 pKa = 11.84RR29 pKa = 11.84DD30 pKa = 3.17GRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84AEE36 pKa = 4.11VIVGNLDD43 pKa = 3.36EE44 pKa = 4.93LVRR47 pKa = 11.84EE48 pKa = 4.23VLGNYY53 pKa = 7.33PARR56 pKa = 11.84RR57 pKa = 11.84EE58 pKa = 4.06ALEE61 pKa = 3.94AAVEE65 pKa = 4.41GWRR68 pKa = 11.84TIPTASRR75 pKa = 11.84RR76 pKa = 11.84VVLRR80 pKa = 11.84HH81 pKa = 4.25GRR83 pKa = 11.84GRR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84SSRR93 pKa = 11.84RR94 pKa = 11.84SGPAAGRR101 pKa = 11.84RR102 pKa = 11.84SSCRR106 pKa = 11.84GRR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84SSRR116 pKa = 11.84RR117 pKa = 11.84RR118 pKa = 11.84QDD120 pKa = 3.63ALLEE124 pKa = 4.28DD125 pKa = 4.93LPDD128 pKa = 4.2EE129 pKa = 5.2GDD131 pKa = 4.15DD132 pKa = 3.98GPSRR136 pKa = 11.84EE137 pKa = 4.15FRR139 pKa = 11.84AAQQDD144 pKa = 4.04GYY146 pKa = 11.15LRR148 pKa = 11.84QGDD151 pKa = 3.85SDD153 pKa = 5.0DD154 pKa = 3.88PDD156 pKa = 3.4EE157 pKa = 5.98RR158 pKa = 11.84EE159 pKa = 4.0GLGQFGARR167 pKa = 11.84RR168 pKa = 11.84SDD170 pKa = 3.44TQALEE175 pKa = 4.66DD176 pKa = 4.43FEE178 pKa = 6.97SDD180 pKa = 3.05AA181 pKa = 5.22
Molecular weight: 20.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4271 |
61 |
482 |
142.4 |
15.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.342 ± 0.828 | 0.913 ± 0.248 |
9.412 ± 0.57 | 8.616 ± 1.029 |
2.81 ± 0.286 | 8.523 ± 0.903 |
1.545 ± 0.252 | 4.121 ± 0.312 |
2.458 ± 0.433 | 8.031 ± 0.548 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.084 ± 0.263 | 2.88 ± 0.612 |
4.893 ± 0.411 | 3.114 ± 0.371 |
7.68 ± 1.075 | 6.392 ± 0.677 |
6.532 ± 0.659 | 6.556 ± 0.519 |
1.616 ± 0.217 | 2.482 ± 0.411 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
