
Gemycircularvirus gemy-ch-rat1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus ratas1; Rat associated gemycircularvirus 1
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
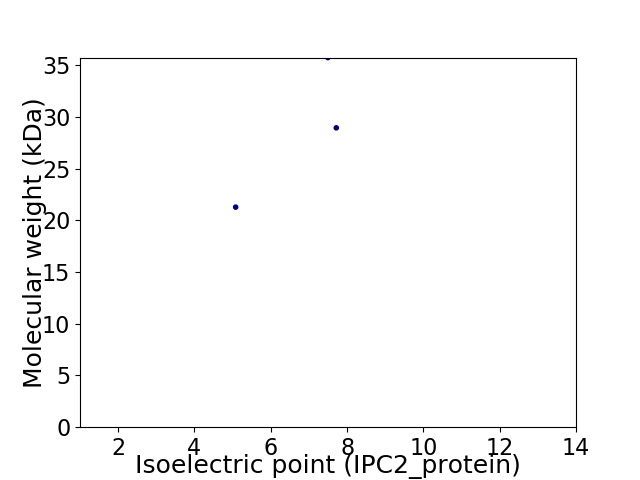
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
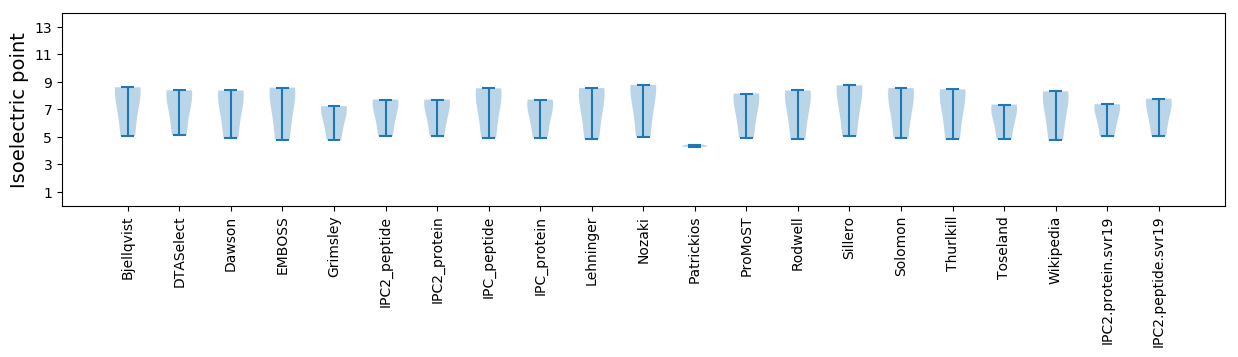
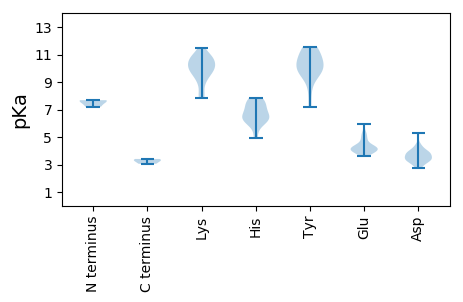
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M5IIA1|A0A0M5IIA1_9VIRU Uncharacterized protein OS=Gemycircularvirus gemy-ch-rat1 OX=1708653 PE=4 SV=1
MM1 pKa = 7.68PSSPMHH7 pKa = 5.94NVGDD11 pKa = 4.0LGFLGLLWGCSQRR24 pKa = 11.84IEE26 pKa = 4.03LSVPLEE32 pKa = 3.98EE33 pKa = 5.21SLTLMGEE40 pKa = 4.48FTSTLSSISADD51 pKa = 3.03SSAVEE56 pKa = 3.62RR57 pKa = 11.84LMFSMWEE64 pKa = 3.97AATLTYY70 pKa = 10.52RR71 pKa = 11.84SLEE74 pKa = 4.0GLQRR78 pKa = 11.84RR79 pKa = 11.84ATTTGSRR86 pKa = 11.84MAMWSQEE93 pKa = 4.02DD94 pKa = 3.85SNGRR98 pKa = 11.84YY99 pKa = 9.49LDD101 pKa = 3.67RR102 pKa = 11.84AEE104 pKa = 3.97VALARR109 pKa = 11.84LVLSGLQLRR118 pKa = 11.84WQRR121 pKa = 11.84TQEE124 pKa = 3.86NFGDD128 pKa = 5.28FVTSWLQNMSAALSTASGATQTGSSGLWMNPTLPPMEE165 pKa = 4.87YY166 pKa = 10.43GLTRR170 pKa = 11.84QRR172 pKa = 11.84LRR174 pKa = 11.84EE175 pKa = 4.14STSGSVRR182 pKa = 11.84LVWDD186 pKa = 4.34LDD188 pKa = 3.31HH189 pKa = 7.47WDD191 pKa = 3.3
MM1 pKa = 7.68PSSPMHH7 pKa = 5.94NVGDD11 pKa = 4.0LGFLGLLWGCSQRR24 pKa = 11.84IEE26 pKa = 4.03LSVPLEE32 pKa = 3.98EE33 pKa = 5.21SLTLMGEE40 pKa = 4.48FTSTLSSISADD51 pKa = 3.03SSAVEE56 pKa = 3.62RR57 pKa = 11.84LMFSMWEE64 pKa = 3.97AATLTYY70 pKa = 10.52RR71 pKa = 11.84SLEE74 pKa = 4.0GLQRR78 pKa = 11.84RR79 pKa = 11.84ATTTGSRR86 pKa = 11.84MAMWSQEE93 pKa = 4.02DD94 pKa = 3.85SNGRR98 pKa = 11.84YY99 pKa = 9.49LDD101 pKa = 3.67RR102 pKa = 11.84AEE104 pKa = 3.97VALARR109 pKa = 11.84LVLSGLQLRR118 pKa = 11.84WQRR121 pKa = 11.84TQEE124 pKa = 3.86NFGDD128 pKa = 5.28FVTSWLQNMSAALSTASGATQTGSSGLWMNPTLPPMEE165 pKa = 4.87YY166 pKa = 10.43GLTRR170 pKa = 11.84QRR172 pKa = 11.84LRR174 pKa = 11.84EE175 pKa = 4.14STSGSVRR182 pKa = 11.84LVWDD186 pKa = 4.34LDD188 pKa = 3.31HH189 pKa = 7.47WDD191 pKa = 3.3
Molecular weight: 21.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M5IIA1|A0A0M5IIA1_9VIRU Uncharacterized protein OS=Gemycircularvirus gemy-ch-rat1 OX=1708653 PE=4 SV=1
MM1 pKa = 7.2SRR3 pKa = 11.84KK4 pKa = 9.85KK5 pKa = 10.26ILNLTSIEE13 pKa = 4.12KK14 pKa = 10.38RR15 pKa = 11.84NTMLQYY21 pKa = 11.51ANTSTSGGSSVTIGPGPALINGGSNGFFILCPTAMDD57 pKa = 3.82LVEE60 pKa = 5.35RR61 pKa = 11.84GGANNSIVNAAEE73 pKa = 4.07RR74 pKa = 11.84NSTTRR79 pKa = 11.84YY80 pKa = 8.38MRR82 pKa = 11.84GFSEE86 pKa = 4.76NIRR89 pKa = 11.84IQTSTGLPWFWRR101 pKa = 11.84RR102 pKa = 11.84ICFCAKK108 pKa = 10.25NSIFNTFFSGDD119 pKa = 3.27TPTPNKK125 pKa = 9.95SSNVSYY131 pKa = 11.56VDD133 pKa = 3.15TTNGMEE139 pKa = 4.18RR140 pKa = 11.84LWFNQSVKK148 pKa = 9.09ITPLTQTAIQSVLFKK163 pKa = 10.89GISGQDD169 pKa = 2.9WTDD172 pKa = 3.18VLTAPVDD179 pKa = 3.43THH181 pKa = 7.61RR182 pKa = 11.84VDD184 pKa = 4.49LKK186 pKa = 10.51SDD188 pKa = 2.89KK189 pKa = 9.87MKK191 pKa = 10.8CIRR194 pKa = 11.84SGNASGAVSEE204 pKa = 4.24RR205 pKa = 11.84KK206 pKa = 9.04YY207 pKa = 9.98WYY209 pKa = 9.7PMNKK213 pKa = 9.54NLVLNDD219 pKa = 4.01DD220 pKa = 3.87EE221 pKa = 5.97SGEE224 pKa = 4.42KK225 pKa = 8.05MTTDD229 pKa = 3.63YY230 pKa = 11.47QSVLDD235 pKa = 3.85KK236 pKa = 11.51QEE238 pKa = 4.23DD239 pKa = 3.41GGYY242 pKa = 10.33YY243 pKa = 9.74IVDD246 pKa = 3.86VFQPGTGATSTDD258 pKa = 3.35QLQVV262 pKa = 3.05
MM1 pKa = 7.2SRR3 pKa = 11.84KK4 pKa = 9.85KK5 pKa = 10.26ILNLTSIEE13 pKa = 4.12KK14 pKa = 10.38RR15 pKa = 11.84NTMLQYY21 pKa = 11.51ANTSTSGGSSVTIGPGPALINGGSNGFFILCPTAMDD57 pKa = 3.82LVEE60 pKa = 5.35RR61 pKa = 11.84GGANNSIVNAAEE73 pKa = 4.07RR74 pKa = 11.84NSTTRR79 pKa = 11.84YY80 pKa = 8.38MRR82 pKa = 11.84GFSEE86 pKa = 4.76NIRR89 pKa = 11.84IQTSTGLPWFWRR101 pKa = 11.84RR102 pKa = 11.84ICFCAKK108 pKa = 10.25NSIFNTFFSGDD119 pKa = 3.27TPTPNKK125 pKa = 9.95SSNVSYY131 pKa = 11.56VDD133 pKa = 3.15TTNGMEE139 pKa = 4.18RR140 pKa = 11.84LWFNQSVKK148 pKa = 9.09ITPLTQTAIQSVLFKK163 pKa = 10.89GISGQDD169 pKa = 2.9WTDD172 pKa = 3.18VLTAPVDD179 pKa = 3.43THH181 pKa = 7.61RR182 pKa = 11.84VDD184 pKa = 4.49LKK186 pKa = 10.51SDD188 pKa = 2.89KK189 pKa = 9.87MKK191 pKa = 10.8CIRR194 pKa = 11.84SGNASGAVSEE204 pKa = 4.24RR205 pKa = 11.84KK206 pKa = 9.04YY207 pKa = 9.98WYY209 pKa = 9.7PMNKK213 pKa = 9.54NLVLNDD219 pKa = 4.01DD220 pKa = 3.87EE221 pKa = 5.97SGEE224 pKa = 4.42KK225 pKa = 8.05MTTDD229 pKa = 3.63YY230 pKa = 11.47QSVLDD235 pKa = 3.85KK236 pKa = 11.51QEE238 pKa = 4.23DD239 pKa = 3.41GGYY242 pKa = 10.33YY243 pKa = 9.74IVDD246 pKa = 3.86VFQPGTGATSTDD258 pKa = 3.35QLQVV262 pKa = 3.05
Molecular weight: 28.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
775 |
191 |
322 |
258.3 |
28.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.968 ± 1.29 | 1.935 ± 0.669 |
5.032 ± 0.357 | 5.032 ± 0.75 |
3.871 ± 0.427 | 9.161 ± 0.776 |
1.935 ± 1.024 | 4.0 ± 1.069 |
3.613 ± 1.25 | 8.129 ± 2.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.097 ± 0.807 | 4.774 ± 1.339 |
3.742 ± 0.214 | 3.613 ± 0.627 |
6.71 ± 0.815 | 9.806 ± 1.485 |
6.968 ± 1.874 | 5.935 ± 0.631 |
2.968 ± 0.535 | 2.71 ± 0.386 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
