
Gordonia phage McGonagall
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
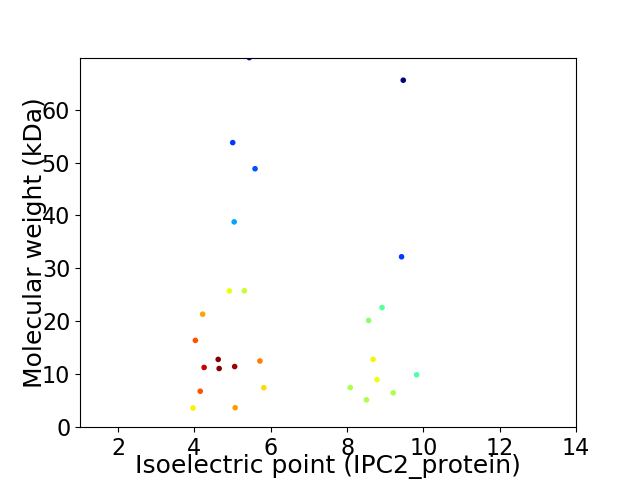
Virtual 2D-PAGE plot for 27 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A160DJQ6|A0A160DJQ6_9CAUD ABC transporter ATP-binding protein OS=Gordonia phage McGonagall OX=1838072 GN=27 PE=4 SV=1
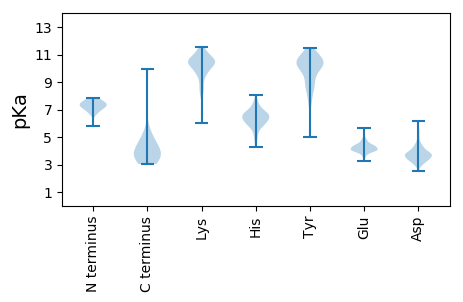
MM1 pKa = 7.84PATTPAPYY9 pKa = 10.22SLPYY13 pKa = 10.58LVTSDD18 pKa = 4.68PLHH21 pKa = 6.69QIAQNTRR28 pKa = 11.84NLAEE32 pKa = 4.33RR33 pKa = 11.84LALLIASGEE42 pKa = 4.26LTGDD46 pKa = 3.58TGPANTLSIGTVTTAPAGDD65 pKa = 3.84PAAATISGIAPNQTLSLTLPRR86 pKa = 11.84GGQGDD91 pKa = 3.88TGPANTLSIGTVTTAPAGDD110 pKa = 3.84PAAATISGSAPNQTLSLTLPQGTGLHH136 pKa = 6.02LLGVVSTVAEE146 pKa = 4.51LGPAGSPGDD155 pKa = 3.77AKK157 pKa = 10.84LVLSDD162 pKa = 5.14PPHH165 pKa = 6.36LWVSDD170 pKa = 3.36GTSWHH175 pKa = 7.67DD176 pKa = 3.36SGQLQGPAGEE186 pKa = 4.36PAPDD190 pKa = 3.4QVTGAGLTLAVSSSPPAAGTPATTITVVSS219 pKa = 3.99
MM1 pKa = 7.84PATTPAPYY9 pKa = 10.22SLPYY13 pKa = 10.58LVTSDD18 pKa = 4.68PLHH21 pKa = 6.69QIAQNTRR28 pKa = 11.84NLAEE32 pKa = 4.33RR33 pKa = 11.84LALLIASGEE42 pKa = 4.26LTGDD46 pKa = 3.58TGPANTLSIGTVTTAPAGDD65 pKa = 3.84PAAATISGIAPNQTLSLTLPRR86 pKa = 11.84GGQGDD91 pKa = 3.88TGPANTLSIGTVTTAPAGDD110 pKa = 3.84PAAATISGSAPNQTLSLTLPQGTGLHH136 pKa = 6.02LLGVVSTVAEE146 pKa = 4.51LGPAGSPGDD155 pKa = 3.77AKK157 pKa = 10.84LVLSDD162 pKa = 5.14PPHH165 pKa = 6.36LWVSDD170 pKa = 3.36GTSWHH175 pKa = 7.67DD176 pKa = 3.36SGQLQGPAGEE186 pKa = 4.36PAPDD190 pKa = 3.4QVTGAGLTLAVSSSPPAAGTPATTITVVSS219 pKa = 3.99
Molecular weight: 21.35 kDa
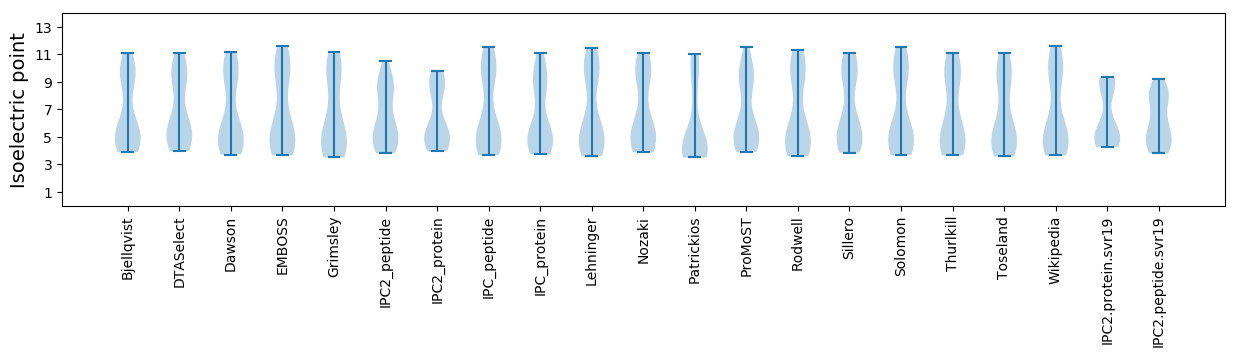
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A160DHC0|A0A160DHC0_9CAUD Uncharacterized protein OS=Gordonia phage McGonagall OX=1838072 GN=12 PE=4 SV=1
MM1 pKa = 6.81VATRR5 pKa = 11.84ARR7 pKa = 11.84FGGDD11 pKa = 3.12VVALAAPVRR20 pKa = 11.84SSDD23 pKa = 3.83DD24 pKa = 3.68LSDD27 pKa = 3.56VLMWSAAIAAFVSYY41 pKa = 10.16LGAGGRR47 pKa = 11.84SLEE50 pKa = 4.17TRR52 pKa = 11.84KK53 pKa = 9.75LRR55 pKa = 11.84QYY57 pKa = 11.27QLLCFADD64 pKa = 3.81VATAAPSAVEE74 pKa = 4.06LPEE77 pKa = 3.87LVAWLDD83 pKa = 3.53HH84 pKa = 6.51EE85 pKa = 4.76GWSNNTRR92 pKa = 11.84RR93 pKa = 11.84SVRR96 pKa = 11.84ACLVVFFTWLQATGQRR112 pKa = 11.84PDD114 pKa = 3.05NPARR118 pKa = 11.84LLPTVAAKK126 pKa = 10.14PGRR129 pKa = 11.84PRR131 pKa = 11.84PCPEE135 pKa = 3.5SALRR139 pKa = 11.84SAVVNAAPRR148 pKa = 11.84EE149 pKa = 3.97RR150 pKa = 11.84LMLALAATGGLRR162 pKa = 11.84RR163 pKa = 11.84AEE165 pKa = 3.78IAAVRR170 pKa = 11.84GEE172 pKa = 4.06HH173 pKa = 5.75VEE175 pKa = 4.03RR176 pKa = 11.84GFDD179 pKa = 3.64GAQLRR184 pKa = 11.84VLGKK188 pKa = 10.06GGRR191 pKa = 11.84EE192 pKa = 3.41RR193 pKa = 11.84LVPISDD199 pKa = 3.7EE200 pKa = 3.99LAEE203 pKa = 3.93QLEE206 pKa = 4.08QRR208 pKa = 11.84RR209 pKa = 11.84GYY211 pKa = 10.91AFPSPRR217 pKa = 11.84GGHH220 pKa = 5.94LTPAHH225 pKa = 5.78VGKK228 pKa = 10.72LIARR232 pKa = 11.84LLPPGWTTHH241 pKa = 4.72TLRR244 pKa = 11.84HH245 pKa = 5.89RR246 pKa = 11.84FASAAYY252 pKa = 9.79RR253 pKa = 11.84ADD255 pKa = 3.86RR256 pKa = 11.84DD257 pKa = 3.24IRR259 pKa = 11.84AVQEE263 pKa = 4.27LLGHH267 pKa = 6.66ASVATTQIYY276 pKa = 7.54TALPDD281 pKa = 3.89DD282 pKa = 4.01SLRR285 pKa = 11.84RR286 pKa = 11.84ATLSATTFAAVAA298 pKa = 3.8
MM1 pKa = 6.81VATRR5 pKa = 11.84ARR7 pKa = 11.84FGGDD11 pKa = 3.12VVALAAPVRR20 pKa = 11.84SSDD23 pKa = 3.83DD24 pKa = 3.68LSDD27 pKa = 3.56VLMWSAAIAAFVSYY41 pKa = 10.16LGAGGRR47 pKa = 11.84SLEE50 pKa = 4.17TRR52 pKa = 11.84KK53 pKa = 9.75LRR55 pKa = 11.84QYY57 pKa = 11.27QLLCFADD64 pKa = 3.81VATAAPSAVEE74 pKa = 4.06LPEE77 pKa = 3.87LVAWLDD83 pKa = 3.53HH84 pKa = 6.51EE85 pKa = 4.76GWSNNTRR92 pKa = 11.84RR93 pKa = 11.84SVRR96 pKa = 11.84ACLVVFFTWLQATGQRR112 pKa = 11.84PDD114 pKa = 3.05NPARR118 pKa = 11.84LLPTVAAKK126 pKa = 10.14PGRR129 pKa = 11.84PRR131 pKa = 11.84PCPEE135 pKa = 3.5SALRR139 pKa = 11.84SAVVNAAPRR148 pKa = 11.84EE149 pKa = 3.97RR150 pKa = 11.84LMLALAATGGLRR162 pKa = 11.84RR163 pKa = 11.84AEE165 pKa = 3.78IAAVRR170 pKa = 11.84GEE172 pKa = 4.06HH173 pKa = 5.75VEE175 pKa = 4.03RR176 pKa = 11.84GFDD179 pKa = 3.64GAQLRR184 pKa = 11.84VLGKK188 pKa = 10.06GGRR191 pKa = 11.84EE192 pKa = 3.41RR193 pKa = 11.84LVPISDD199 pKa = 3.7EE200 pKa = 3.99LAEE203 pKa = 3.93QLEE206 pKa = 4.08QRR208 pKa = 11.84RR209 pKa = 11.84GYY211 pKa = 10.91AFPSPRR217 pKa = 11.84GGHH220 pKa = 5.94LTPAHH225 pKa = 5.78VGKK228 pKa = 10.72LIARR232 pKa = 11.84LLPPGWTTHH241 pKa = 4.72TLRR244 pKa = 11.84HH245 pKa = 5.89RR246 pKa = 11.84FASAAYY252 pKa = 9.79RR253 pKa = 11.84ADD255 pKa = 3.86RR256 pKa = 11.84DD257 pKa = 3.24IRR259 pKa = 11.84AVQEE263 pKa = 4.27LLGHH267 pKa = 6.66ASVATTQIYY276 pKa = 7.54TALPDD281 pKa = 3.89DD282 pKa = 4.01SLRR285 pKa = 11.84RR286 pKa = 11.84ATLSATTFAAVAA298 pKa = 3.8
Molecular weight: 32.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5413 |
33 |
657 |
200.5 |
21.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.576 ± 0.806 | 0.887 ± 0.202 |
6.263 ± 0.649 | 3.769 ± 0.439 |
2.476 ± 0.203 | 8.406 ± 0.72 |
2.161 ± 0.301 | 4.194 ± 0.607 |
2.346 ± 0.256 | 8.72 ± 0.426 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.014 ± 0.175 | 2.512 ± 0.285 |
6.484 ± 0.474 | 2.937 ± 0.232 |
6.392 ± 0.713 | 6.281 ± 0.374 |
7.741 ± 0.448 | 8.239 ± 0.6 |
1.774 ± 0.121 | 1.829 ± 0.258 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
