
Pseudidiomarina aestuarii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Idiomarinaceae; Pseudidiomarina
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
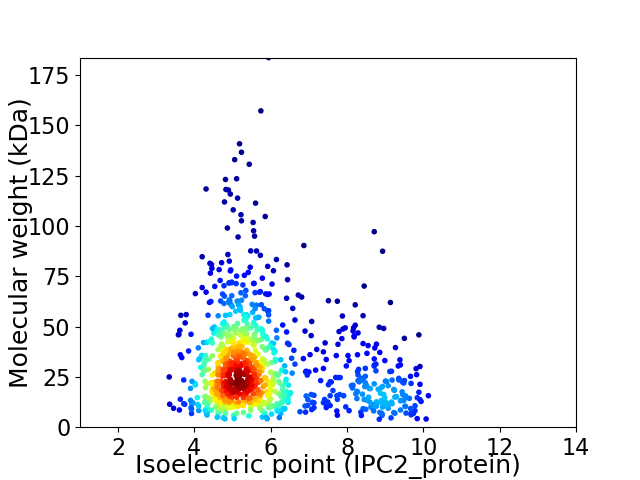
Virtual 2D-PAGE plot for 947 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T4D981|A0A2T4D981_9GAMM Cellulose synthase catalytic subunit [UDP-forming] OS=Pseudidiomarina aestuarii OX=624146 GN=bcsA PE=3 SV=1
MM1 pKa = 7.12TAGLRR6 pKa = 11.84YY7 pKa = 9.34LAILSLAFTLAACGSGEE24 pKa = 4.21RR25 pKa = 11.84NTGGGDD31 pKa = 3.08TGGDD35 pKa = 3.45GGGSGSTPPTATASNVTVNEE55 pKa = 4.1GNPNQVIEE63 pKa = 4.09AAITVSLSTTANASVSFDD81 pKa = 3.61YY82 pKa = 9.48ATADD86 pKa = 3.23ASGLAGTDD94 pKa = 3.64YY95 pKa = 11.1EE96 pKa = 4.52DD97 pKa = 3.5TSGTVTFTAGQSSQTITIPIISNAVFNDD125 pKa = 3.11DD126 pKa = 3.68RR127 pKa = 11.84DD128 pKa = 4.04FFVDD132 pKa = 4.54FSNAQGLTLGTTRR145 pKa = 11.84VQVTIVNDD153 pKa = 3.59DD154 pKa = 3.77PRR156 pKa = 11.84PVLAFEE162 pKa = 3.9QSIFEE167 pKa = 4.35TTEE170 pKa = 3.56QAGTLGLTVTLSNGTEE186 pKa = 4.08SEE188 pKa = 4.46VTFDD192 pKa = 4.62LDD194 pKa = 3.8VSGTATAGVDD204 pKa = 3.51YY205 pKa = 10.77SLEE208 pKa = 4.25EE209 pKa = 3.87TSFTIPAGALEE220 pKa = 4.09QQISLTLIADD230 pKa = 3.8DD231 pKa = 3.82VKK233 pKa = 11.23EE234 pKa = 4.36GGEE237 pKa = 4.31SVILAFANVRR247 pKa = 11.84EE248 pKa = 4.3ADD250 pKa = 3.94TNPDD254 pKa = 3.57LSTQTIVIRR263 pKa = 11.84GDD265 pKa = 3.79AILADD270 pKa = 3.92TGAVEE275 pKa = 4.8YY276 pKa = 10.81YY277 pKa = 10.82NSGDD281 pKa = 3.58FSAATPDD288 pKa = 3.96AEE290 pKa = 5.54HH291 pKa = 7.35PNQDD295 pKa = 2.74ADD297 pKa = 3.76YY298 pKa = 11.23GRR300 pKa = 11.84DD301 pKa = 2.94TTAAVDD307 pKa = 3.81NNADD311 pKa = 3.43GAGNLNLTKK320 pKa = 10.3LDD322 pKa = 3.54SSGNALPQNATSFSCVRR339 pKa = 11.84DD340 pKa = 3.78NLSGTVYY347 pKa = 10.75AVWAEE352 pKa = 3.8GDD354 pKa = 3.67ALSNFYY360 pKa = 10.91FNPTYY365 pKa = 10.21RR366 pKa = 11.84YY367 pKa = 9.81KK368 pKa = 10.55WFNDD372 pKa = 3.11DD373 pKa = 3.44STNNAGNPGTFTPEE387 pKa = 3.62SAFPAFPDD395 pKa = 3.62EE396 pKa = 4.51NDD398 pKa = 3.94PIYY401 pKa = 10.94YY402 pKa = 10.4EE403 pKa = 4.14NANCAFPAVDD413 pKa = 4.32PVVQTLGCSSQNYY426 pKa = 7.87TNYY429 pKa = 10.53LNTIGYY435 pKa = 9.79CGFDD439 pKa = 3.87DD440 pKa = 3.63WRR442 pKa = 11.84LPTISEE448 pKa = 4.17LQSIAIYY455 pKa = 10.27QADD458 pKa = 3.62QSQDD462 pKa = 3.05NLFFPDD468 pKa = 3.47VAMMTPNGDD477 pKa = 3.21GTVRR481 pKa = 11.84LLSSTPAADD490 pKa = 3.98PNSSAWCLEE499 pKa = 3.89VQTARR504 pKa = 11.84RR505 pKa = 11.84MLCNKK510 pKa = 9.85NDD512 pKa = 3.61VNSIRR517 pKa = 11.84AARR520 pKa = 11.84NPVAYY525 pKa = 10.27QEE527 pKa = 4.18
MM1 pKa = 7.12TAGLRR6 pKa = 11.84YY7 pKa = 9.34LAILSLAFTLAACGSGEE24 pKa = 4.21RR25 pKa = 11.84NTGGGDD31 pKa = 3.08TGGDD35 pKa = 3.45GGGSGSTPPTATASNVTVNEE55 pKa = 4.1GNPNQVIEE63 pKa = 4.09AAITVSLSTTANASVSFDD81 pKa = 3.61YY82 pKa = 9.48ATADD86 pKa = 3.23ASGLAGTDD94 pKa = 3.64YY95 pKa = 11.1EE96 pKa = 4.52DD97 pKa = 3.5TSGTVTFTAGQSSQTITIPIISNAVFNDD125 pKa = 3.11DD126 pKa = 3.68RR127 pKa = 11.84DD128 pKa = 4.04FFVDD132 pKa = 4.54FSNAQGLTLGTTRR145 pKa = 11.84VQVTIVNDD153 pKa = 3.59DD154 pKa = 3.77PRR156 pKa = 11.84PVLAFEE162 pKa = 3.9QSIFEE167 pKa = 4.35TTEE170 pKa = 3.56QAGTLGLTVTLSNGTEE186 pKa = 4.08SEE188 pKa = 4.46VTFDD192 pKa = 4.62LDD194 pKa = 3.8VSGTATAGVDD204 pKa = 3.51YY205 pKa = 10.77SLEE208 pKa = 4.25EE209 pKa = 3.87TSFTIPAGALEE220 pKa = 4.09QQISLTLIADD230 pKa = 3.8DD231 pKa = 3.82VKK233 pKa = 11.23EE234 pKa = 4.36GGEE237 pKa = 4.31SVILAFANVRR247 pKa = 11.84EE248 pKa = 4.3ADD250 pKa = 3.94TNPDD254 pKa = 3.57LSTQTIVIRR263 pKa = 11.84GDD265 pKa = 3.79AILADD270 pKa = 3.92TGAVEE275 pKa = 4.8YY276 pKa = 10.81YY277 pKa = 10.82NSGDD281 pKa = 3.58FSAATPDD288 pKa = 3.96AEE290 pKa = 5.54HH291 pKa = 7.35PNQDD295 pKa = 2.74ADD297 pKa = 3.76YY298 pKa = 11.23GRR300 pKa = 11.84DD301 pKa = 2.94TTAAVDD307 pKa = 3.81NNADD311 pKa = 3.43GAGNLNLTKK320 pKa = 10.3LDD322 pKa = 3.54SSGNALPQNATSFSCVRR339 pKa = 11.84DD340 pKa = 3.78NLSGTVYY347 pKa = 10.75AVWAEE352 pKa = 3.8GDD354 pKa = 3.67ALSNFYY360 pKa = 10.91FNPTYY365 pKa = 10.21RR366 pKa = 11.84YY367 pKa = 9.81KK368 pKa = 10.55WFNDD372 pKa = 3.11DD373 pKa = 3.44STNNAGNPGTFTPEE387 pKa = 3.62SAFPAFPDD395 pKa = 3.62EE396 pKa = 4.51NDD398 pKa = 3.94PIYY401 pKa = 10.94YY402 pKa = 10.4EE403 pKa = 4.14NANCAFPAVDD413 pKa = 4.32PVVQTLGCSSQNYY426 pKa = 7.87TNYY429 pKa = 10.53LNTIGYY435 pKa = 9.79CGFDD439 pKa = 3.87DD440 pKa = 3.63WRR442 pKa = 11.84LPTISEE448 pKa = 4.17LQSIAIYY455 pKa = 10.27QADD458 pKa = 3.62QSQDD462 pKa = 3.05NLFFPDD468 pKa = 3.47VAMMTPNGDD477 pKa = 3.21GTVRR481 pKa = 11.84LLSSTPAADD490 pKa = 3.98PNSSAWCLEE499 pKa = 3.89VQTARR504 pKa = 11.84RR505 pKa = 11.84MLCNKK510 pKa = 9.85NDD512 pKa = 3.61VNSIRR517 pKa = 11.84AARR520 pKa = 11.84NPVAYY525 pKa = 10.27QEE527 pKa = 4.18
Molecular weight: 55.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T4CVT6|A0A2T4CVT6_9GAMM Thioesterase family protein OS=Pseudidiomarina aestuarii OX=624146 GN=C9927_02105 PE=4 SV=1
MM1 pKa = 7.42KK2 pKa = 10.05RR3 pKa = 11.84QKK5 pKa = 10.3RR6 pKa = 11.84DD7 pKa = 3.2RR8 pKa = 11.84LEE10 pKa = 3.91RR11 pKa = 11.84AHH13 pKa = 6.72AQGFKK18 pKa = 10.51AGVSGRR24 pKa = 11.84SKK26 pKa = 10.43EE27 pKa = 3.87LCPYY31 pKa = 9.75QNQDD35 pKa = 5.21ARR37 pKa = 11.84SQWLGGWRR45 pKa = 11.84DD46 pKa = 3.18AMEE49 pKa = 4.08ARR51 pKa = 11.84GVGLYY56 pKa = 9.9RR57 pKa = 5.07
MM1 pKa = 7.42KK2 pKa = 10.05RR3 pKa = 11.84QKK5 pKa = 10.3RR6 pKa = 11.84DD7 pKa = 3.2RR8 pKa = 11.84LEE10 pKa = 3.91RR11 pKa = 11.84AHH13 pKa = 6.72AQGFKK18 pKa = 10.51AGVSGRR24 pKa = 11.84SKK26 pKa = 10.43EE27 pKa = 3.87LCPYY31 pKa = 9.75QNQDD35 pKa = 5.21ARR37 pKa = 11.84SQWLGGWRR45 pKa = 11.84DD46 pKa = 3.18AMEE49 pKa = 4.08ARR51 pKa = 11.84GVGLYY56 pKa = 9.9RR57 pKa = 5.07
Molecular weight: 6.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
274386 |
33 |
1619 |
289.7 |
32.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.898 ± 0.084 | 0.836 ± 0.026 |
5.657 ± 0.063 | 6.399 ± 0.083 |
3.997 ± 0.061 | 6.988 ± 0.085 |
2.146 ± 0.042 | 5.801 ± 0.073 |
3.868 ± 0.069 | 10.611 ± 0.081 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.394 ± 0.039 | 3.501 ± 0.051 |
4.248 ± 0.048 | 5.044 ± 0.078 |
5.678 ± 0.07 | 6.013 ± 0.064 |
5.374 ± 0.056 | 7.261 ± 0.075 |
1.36 ± 0.035 | 2.924 ± 0.043 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
