
Lachnospiraceae bacterium M18-1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; unclassified Lachnospiraceae
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
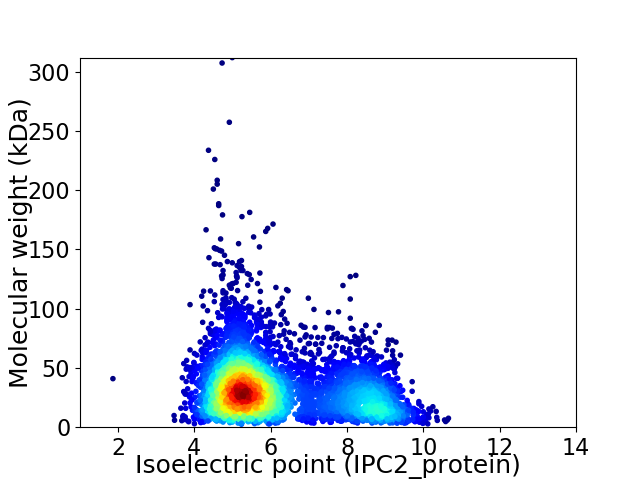
Virtual 2D-PAGE plot for 5263 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9JRF0|R9JRF0_9FIRM ABC transporter domain-containing protein OS=Lachnospiraceae bacterium M18-1 OX=1235792 GN=C808_03980 PE=4 SV=1
MM1 pKa = 7.49KK2 pKa = 10.43KK3 pKa = 10.23KK4 pKa = 10.47LLSVLLAGVMVISLAACGGGSGDD27 pKa = 4.19GGSGDD32 pKa = 3.79SDD34 pKa = 3.44GGSGTTAEE42 pKa = 4.58SDD44 pKa = 4.16DD45 pKa = 4.03GGADD49 pKa = 3.27SGSEE53 pKa = 4.1GGHH56 pKa = 6.77KK57 pKa = 9.42LTVWTWDD64 pKa = 3.35PAFNIPAIKK73 pKa = 9.92EE74 pKa = 3.63AGEE77 pKa = 4.0IYY79 pKa = 10.15KK80 pKa = 10.72AEE82 pKa = 3.95VDD84 pKa = 3.93SEE86 pKa = 4.59FEE88 pKa = 4.26LEE90 pKa = 4.38VIEE93 pKa = 4.72TLSDD97 pKa = 3.54DD98 pKa = 5.17CEE100 pKa = 4.58TKK102 pKa = 10.79LQTCAEE108 pKa = 4.23SGDD111 pKa = 4.07FGTMADD117 pKa = 4.39IILMQDD123 pKa = 2.56NSFQKK128 pKa = 10.27FVSFYY133 pKa = 10.6PEE135 pKa = 4.17AFTDD139 pKa = 3.89LTDD142 pKa = 3.44SGIEE146 pKa = 3.72FSEE149 pKa = 4.36FAEE152 pKa = 4.86GKK154 pKa = 9.59LAYY157 pKa = 9.46STVDD161 pKa = 3.04GKK163 pKa = 10.87NYY165 pKa = 10.06GVPFDD170 pKa = 3.85NGAVISCYY178 pKa = 9.61RR179 pKa = 11.84TDD181 pKa = 3.79ILDD184 pKa = 3.5QAGYY188 pKa = 8.83TIDD191 pKa = 5.13DD192 pKa = 4.21LTDD195 pKa = 3.61IDD197 pKa = 3.54WDD199 pKa = 3.72KK200 pKa = 11.18FIEE203 pKa = 4.07IGKK206 pKa = 6.48TVKK209 pKa = 10.41EE210 pKa = 4.0KK211 pKa = 9.64TGKK214 pKa = 10.83YY215 pKa = 8.69MLSGQANSQDD225 pKa = 3.93IIMMMLQSAGASLFNEE241 pKa = 4.74DD242 pKa = 3.54GTPNLVGNDD251 pKa = 3.88ALNEE255 pKa = 4.55CIDD258 pKa = 4.09IYY260 pKa = 11.35QKK262 pKa = 9.38MVKK265 pKa = 9.96EE266 pKa = 4.57GIFYY270 pKa = 10.12EE271 pKa = 4.28VNEE274 pKa = 3.76WDD276 pKa = 4.43EE277 pKa = 4.41YY278 pKa = 11.13VSSITNEE285 pKa = 3.71QCAGTINGNWIMATIMGMEE304 pKa = 4.25DD305 pKa = 3.2TSGNWAITNMPKK317 pKa = 10.66LMNASGATNYY327 pKa = 10.59SNNGGSSWYY336 pKa = 9.79ISSNCQNTDD345 pKa = 3.18LAFDD349 pKa = 4.06FMAKK353 pKa = 8.94TFAGSVTLYY362 pKa = 11.03EE363 pKa = 4.79NILPTTGAISTWGPAGDD380 pKa = 3.95SEE382 pKa = 5.92AYY384 pKa = 9.2QAPQEE389 pKa = 4.24FWSDD393 pKa = 3.39QPIFATVVEE402 pKa = 4.99FAEE405 pKa = 4.47KK406 pKa = 9.85TPSNNTSAYY415 pKa = 8.36YY416 pKa = 9.76YY417 pKa = 10.16DD418 pKa = 3.54VRR420 pKa = 11.84EE421 pKa = 4.41VISTAIQDD429 pKa = 3.43IMSGGGEE436 pKa = 3.67KK437 pKa = 10.53DD438 pKa = 3.8AILEE442 pKa = 3.99QAQADD447 pKa = 3.83AEE449 pKa = 4.47FYY451 pKa = 10.13YY452 pKa = 10.68QQ453 pKa = 4.31
MM1 pKa = 7.49KK2 pKa = 10.43KK3 pKa = 10.23KK4 pKa = 10.47LLSVLLAGVMVISLAACGGGSGDD27 pKa = 4.19GGSGDD32 pKa = 3.79SDD34 pKa = 3.44GGSGTTAEE42 pKa = 4.58SDD44 pKa = 4.16DD45 pKa = 4.03GGADD49 pKa = 3.27SGSEE53 pKa = 4.1GGHH56 pKa = 6.77KK57 pKa = 9.42LTVWTWDD64 pKa = 3.35PAFNIPAIKK73 pKa = 9.92EE74 pKa = 3.63AGEE77 pKa = 4.0IYY79 pKa = 10.15KK80 pKa = 10.72AEE82 pKa = 3.95VDD84 pKa = 3.93SEE86 pKa = 4.59FEE88 pKa = 4.26LEE90 pKa = 4.38VIEE93 pKa = 4.72TLSDD97 pKa = 3.54DD98 pKa = 5.17CEE100 pKa = 4.58TKK102 pKa = 10.79LQTCAEE108 pKa = 4.23SGDD111 pKa = 4.07FGTMADD117 pKa = 4.39IILMQDD123 pKa = 2.56NSFQKK128 pKa = 10.27FVSFYY133 pKa = 10.6PEE135 pKa = 4.17AFTDD139 pKa = 3.89LTDD142 pKa = 3.44SGIEE146 pKa = 3.72FSEE149 pKa = 4.36FAEE152 pKa = 4.86GKK154 pKa = 9.59LAYY157 pKa = 9.46STVDD161 pKa = 3.04GKK163 pKa = 10.87NYY165 pKa = 10.06GVPFDD170 pKa = 3.85NGAVISCYY178 pKa = 9.61RR179 pKa = 11.84TDD181 pKa = 3.79ILDD184 pKa = 3.5QAGYY188 pKa = 8.83TIDD191 pKa = 5.13DD192 pKa = 4.21LTDD195 pKa = 3.61IDD197 pKa = 3.54WDD199 pKa = 3.72KK200 pKa = 11.18FIEE203 pKa = 4.07IGKK206 pKa = 6.48TVKK209 pKa = 10.41EE210 pKa = 4.0KK211 pKa = 9.64TGKK214 pKa = 10.83YY215 pKa = 8.69MLSGQANSQDD225 pKa = 3.93IIMMMLQSAGASLFNEE241 pKa = 4.74DD242 pKa = 3.54GTPNLVGNDD251 pKa = 3.88ALNEE255 pKa = 4.55CIDD258 pKa = 4.09IYY260 pKa = 11.35QKK262 pKa = 9.38MVKK265 pKa = 9.96EE266 pKa = 4.57GIFYY270 pKa = 10.12EE271 pKa = 4.28VNEE274 pKa = 3.76WDD276 pKa = 4.43EE277 pKa = 4.41YY278 pKa = 11.13VSSITNEE285 pKa = 3.71QCAGTINGNWIMATIMGMEE304 pKa = 4.25DD305 pKa = 3.2TSGNWAITNMPKK317 pKa = 10.66LMNASGATNYY327 pKa = 10.59SNNGGSSWYY336 pKa = 9.79ISSNCQNTDD345 pKa = 3.18LAFDD349 pKa = 4.06FMAKK353 pKa = 8.94TFAGSVTLYY362 pKa = 11.03EE363 pKa = 4.79NILPTTGAISTWGPAGDD380 pKa = 3.95SEE382 pKa = 5.92AYY384 pKa = 9.2QAPQEE389 pKa = 4.24FWSDD393 pKa = 3.39QPIFATVVEE402 pKa = 4.99FAEE405 pKa = 4.47KK406 pKa = 9.85TPSNNTSAYY415 pKa = 8.36YY416 pKa = 9.76YY417 pKa = 10.16DD418 pKa = 3.54VRR420 pKa = 11.84EE421 pKa = 4.41VISTAIQDD429 pKa = 3.43IMSGGGEE436 pKa = 3.67KK437 pKa = 10.53DD438 pKa = 3.8AILEE442 pKa = 3.99QAQADD447 pKa = 3.83AEE449 pKa = 4.47FYY451 pKa = 10.13YY452 pKa = 10.68QQ453 pKa = 4.31
Molecular weight: 48.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9K6U7|R9K6U7_9FIRM Hydroxyethylthiazole kinase OS=Lachnospiraceae bacterium M18-1 OX=1235792 GN=thiM PE=3 SV=1
MM1 pKa = 7.19NEE3 pKa = 3.78VMKK6 pKa = 10.88SLLSHH11 pKa = 5.57QSIRR15 pKa = 11.84AYY17 pKa = 8.76TEE19 pKa = 3.65QMVEE23 pKa = 4.01EE24 pKa = 4.26EE25 pKa = 5.18KK26 pKa = 10.76INQIIQAVQCRR37 pKa = 11.84QRR39 pKa = 11.84RR40 pKa = 11.84TGTTVQEE47 pKa = 4.05NGRR50 pKa = 11.84TLRR53 pKa = 11.84RR54 pKa = 11.84IFISIHH60 pKa = 3.69MTITRR65 pKa = 11.84KK66 pKa = 8.98FRR68 pKa = 11.84KK69 pKa = 9.5CC70 pKa = 3.14
MM1 pKa = 7.19NEE3 pKa = 3.78VMKK6 pKa = 10.88SLLSHH11 pKa = 5.57QSIRR15 pKa = 11.84AYY17 pKa = 8.76TEE19 pKa = 3.65QMVEE23 pKa = 4.01EE24 pKa = 4.26EE25 pKa = 5.18KK26 pKa = 10.76INQIIQAVQCRR37 pKa = 11.84QRR39 pKa = 11.84RR40 pKa = 11.84TGTTVQEE47 pKa = 4.05NGRR50 pKa = 11.84TLRR53 pKa = 11.84RR54 pKa = 11.84IFISIHH60 pKa = 3.69MTITRR65 pKa = 11.84KK66 pKa = 8.98FRR68 pKa = 11.84KK69 pKa = 9.5CC70 pKa = 3.14
Molecular weight: 8.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1644781 |
23 |
2742 |
312.5 |
35.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.463 ± 0.034 | 1.562 ± 0.015 |
5.527 ± 0.023 | 8.089 ± 0.038 |
4.124 ± 0.025 | 7.144 ± 0.031 |
1.81 ± 0.013 | 7.014 ± 0.034 |
6.527 ± 0.032 | 9.008 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.152 ± 0.017 | 4.036 ± 0.021 |
3.387 ± 0.021 | 3.51 ± 0.019 |
5.075 ± 0.028 | 5.766 ± 0.028 |
5.089 ± 0.027 | 6.461 ± 0.031 |
1.017 ± 0.011 | 4.241 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
