
Phocoena phocoena papillomavirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyopipapillomavirus; Dyopipapillomavirus 1
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
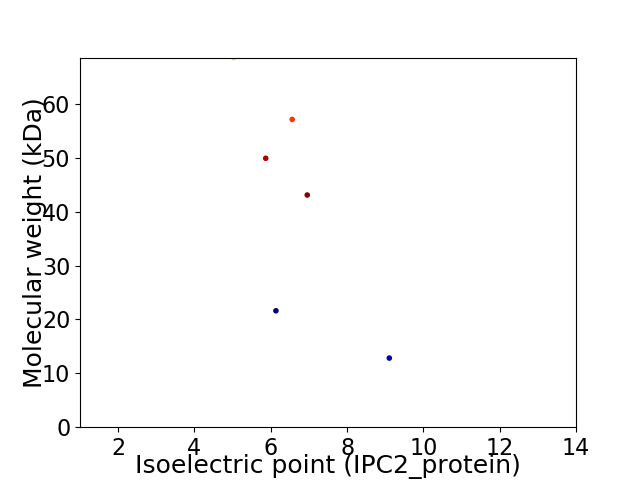
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
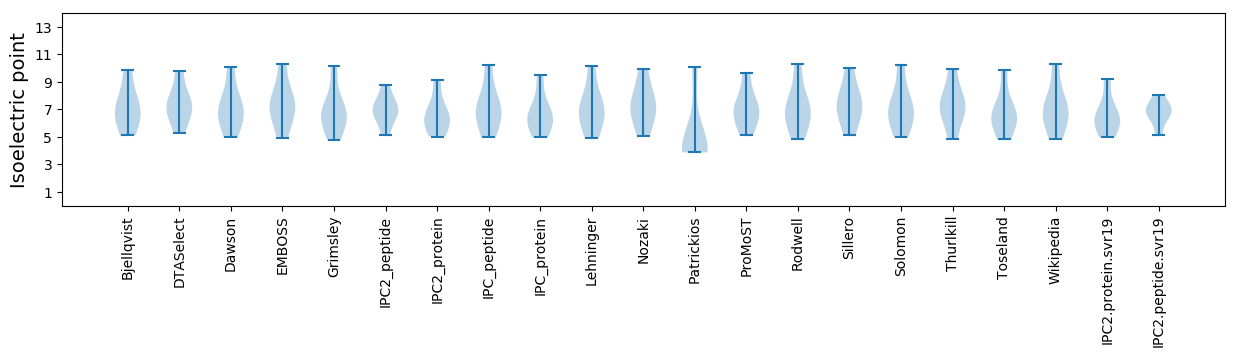
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F2VIS2|F2VIS2_9PAPI Regulatory protein E2 OS=Phocoena phocoena papillomavirus 4 OX=706527 GN=E2 PE=3 SV=1
MM1 pKa = 8.15DD2 pKa = 5.89PPQGTDD8 pKa = 3.02GDD10 pKa = 4.34EE11 pKa = 4.89GCSGWCFVDD20 pKa = 6.23LEE22 pKa = 4.2AQEE25 pKa = 5.81DD26 pKa = 4.67GVDD29 pKa = 5.04DD30 pKa = 4.67NDD32 pKa = 4.38NDD34 pKa = 4.13DD35 pKa = 3.92EE36 pKa = 4.95SVDD39 pKa = 3.71EE40 pKa = 4.07EE41 pKa = 5.05DD42 pKa = 3.77YY43 pKa = 11.51AIVNLFDD50 pKa = 6.13DD51 pKa = 4.43EE52 pKa = 6.34DD53 pKa = 4.01VTQGNSLTVYY63 pKa = 9.31EE64 pKa = 4.38AQNRR68 pKa = 11.84ADD70 pKa = 4.87DD71 pKa = 3.73EE72 pKa = 4.51RR73 pKa = 11.84AVQKK77 pKa = 10.6LKK79 pKa = 10.9RR80 pKa = 11.84KK81 pKa = 9.15LLQSPEE87 pKa = 4.44LDD89 pKa = 3.15LSPRR93 pKa = 11.84LSAITIGKK101 pKa = 9.97KK102 pKa = 9.39SGKK105 pKa = 9.46AKK107 pKa = 10.15RR108 pKa = 11.84RR109 pKa = 11.84LFMPGQDD116 pKa = 2.89SGYY119 pKa = 8.6GQSVEE124 pKa = 4.32ANVEE128 pKa = 4.03EE129 pKa = 5.32SVLQTQVSLNFNGEE143 pKa = 3.96DD144 pKa = 3.4SGGGGGAPCTVSGAQEE160 pKa = 4.14TPEE163 pKa = 4.56TMIQLLKK170 pKa = 10.18QANPQAAMMGLFKK183 pKa = 10.26KK184 pKa = 9.36TMGISFTDD192 pKa = 3.91LSRR195 pKa = 11.84TFQSDD200 pKa = 3.17KK201 pKa = 10.33TPCVDD206 pKa = 3.14WVAVCFGVPLSLIDD220 pKa = 3.58ACTEE224 pKa = 3.77LLQPHH229 pKa = 6.63CEE231 pKa = 4.06FGNVTTTPCCTGYY244 pKa = 11.2LLLLLARR251 pKa = 11.84FQVAKK256 pKa = 10.66CRR258 pKa = 11.84VTVKK262 pKa = 10.69KK263 pKa = 10.85LLGNTLHH270 pKa = 7.23VPEE273 pKa = 4.24QQILTEE279 pKa = 4.19PPKK282 pKa = 10.33IRR284 pKa = 11.84CPAAAIYY291 pKa = 8.97WYY293 pKa = 10.25KK294 pKa = 10.89KK295 pKa = 10.39GITGVAKK302 pKa = 9.33TWGPLPEE309 pKa = 4.34WLAKK313 pKa = 8.54MVNVSSNLADD323 pKa = 3.25AAVFNLSDD331 pKa = 4.35MIQWAYY337 pKa = 11.19DD338 pKa = 3.45NNKK341 pKa = 8.75TEE343 pKa = 4.93DD344 pKa = 3.27ADD346 pKa = 3.39IAYY349 pKa = 9.74GYY351 pKa = 10.47AQLADD356 pKa = 3.43TDD358 pKa = 4.25RR359 pKa = 11.84NAEE362 pKa = 4.12AFLKK366 pKa = 10.95SNSQAKK372 pKa = 8.57YY373 pKa = 10.44VKK375 pKa = 10.13DD376 pKa = 3.53CGVMVRR382 pKa = 11.84LYY384 pKa = 10.73KK385 pKa = 10.13KK386 pKa = 10.81AEE388 pKa = 3.91MQRR391 pKa = 11.84TSMKK395 pKa = 10.27DD396 pKa = 2.91WIKK399 pKa = 10.62RR400 pKa = 11.84RR401 pKa = 11.84CEE403 pKa = 3.93EE404 pKa = 4.07VDD406 pKa = 3.84GDD408 pKa = 4.32GDD410 pKa = 3.39WRR412 pKa = 11.84PIVSFLKK419 pKa = 10.72FQGIDD424 pKa = 3.06MAVFISVLKK433 pKa = 10.77NFLKK437 pKa = 10.92GIPKK441 pKa = 9.61QNCIVICGPPNTGKK455 pKa = 10.6SLFSVSLIQFLSGKK469 pKa = 10.0VISFVNSKK477 pKa = 8.57SHH479 pKa = 7.02FWLQPLGEE487 pKa = 4.64CKK489 pKa = 10.17VALLDD494 pKa = 5.24DD495 pKa = 4.1ATGPAWDD502 pKa = 4.22YY503 pKa = 11.58FDD505 pKa = 3.62TFLRR509 pKa = 11.84NLVDD513 pKa = 4.08GNPISLDD520 pKa = 3.89CKK522 pKa = 10.46HH523 pKa = 6.62KK524 pKa = 11.09APTQVKK530 pKa = 9.21CPPLIITSNIDD541 pKa = 2.95VTASDD546 pKa = 3.02RR547 pKa = 11.84WKK549 pKa = 10.49YY550 pKa = 10.0LHH552 pKa = 7.01SRR554 pKa = 11.84LKK556 pKa = 11.04LFTFSNPCPLTEE568 pKa = 4.04QGEE571 pKa = 4.37PAFNLTPVNWKK582 pKa = 10.37CFFKK586 pKa = 10.76RR587 pKa = 11.84LKK589 pKa = 9.68TRR591 pKa = 11.84LGLADD596 pKa = 5.7DD597 pKa = 4.35EE598 pKa = 5.48GEE600 pKa = 4.18DD601 pKa = 4.1HH602 pKa = 7.37GEE604 pKa = 3.89SQQPLKK610 pKa = 10.17CTARR614 pKa = 11.84GTDD617 pKa = 3.33GFVV620 pKa = 2.77
MM1 pKa = 8.15DD2 pKa = 5.89PPQGTDD8 pKa = 3.02GDD10 pKa = 4.34EE11 pKa = 4.89GCSGWCFVDD20 pKa = 6.23LEE22 pKa = 4.2AQEE25 pKa = 5.81DD26 pKa = 4.67GVDD29 pKa = 5.04DD30 pKa = 4.67NDD32 pKa = 4.38NDD34 pKa = 4.13DD35 pKa = 3.92EE36 pKa = 4.95SVDD39 pKa = 3.71EE40 pKa = 4.07EE41 pKa = 5.05DD42 pKa = 3.77YY43 pKa = 11.51AIVNLFDD50 pKa = 6.13DD51 pKa = 4.43EE52 pKa = 6.34DD53 pKa = 4.01VTQGNSLTVYY63 pKa = 9.31EE64 pKa = 4.38AQNRR68 pKa = 11.84ADD70 pKa = 4.87DD71 pKa = 3.73EE72 pKa = 4.51RR73 pKa = 11.84AVQKK77 pKa = 10.6LKK79 pKa = 10.9RR80 pKa = 11.84KK81 pKa = 9.15LLQSPEE87 pKa = 4.44LDD89 pKa = 3.15LSPRR93 pKa = 11.84LSAITIGKK101 pKa = 9.97KK102 pKa = 9.39SGKK105 pKa = 9.46AKK107 pKa = 10.15RR108 pKa = 11.84RR109 pKa = 11.84LFMPGQDD116 pKa = 2.89SGYY119 pKa = 8.6GQSVEE124 pKa = 4.32ANVEE128 pKa = 4.03EE129 pKa = 5.32SVLQTQVSLNFNGEE143 pKa = 3.96DD144 pKa = 3.4SGGGGGAPCTVSGAQEE160 pKa = 4.14TPEE163 pKa = 4.56TMIQLLKK170 pKa = 10.18QANPQAAMMGLFKK183 pKa = 10.26KK184 pKa = 9.36TMGISFTDD192 pKa = 3.91LSRR195 pKa = 11.84TFQSDD200 pKa = 3.17KK201 pKa = 10.33TPCVDD206 pKa = 3.14WVAVCFGVPLSLIDD220 pKa = 3.58ACTEE224 pKa = 3.77LLQPHH229 pKa = 6.63CEE231 pKa = 4.06FGNVTTTPCCTGYY244 pKa = 11.2LLLLLARR251 pKa = 11.84FQVAKK256 pKa = 10.66CRR258 pKa = 11.84VTVKK262 pKa = 10.69KK263 pKa = 10.85LLGNTLHH270 pKa = 7.23VPEE273 pKa = 4.24QQILTEE279 pKa = 4.19PPKK282 pKa = 10.33IRR284 pKa = 11.84CPAAAIYY291 pKa = 8.97WYY293 pKa = 10.25KK294 pKa = 10.89KK295 pKa = 10.39GITGVAKK302 pKa = 9.33TWGPLPEE309 pKa = 4.34WLAKK313 pKa = 8.54MVNVSSNLADD323 pKa = 3.25AAVFNLSDD331 pKa = 4.35MIQWAYY337 pKa = 11.19DD338 pKa = 3.45NNKK341 pKa = 8.75TEE343 pKa = 4.93DD344 pKa = 3.27ADD346 pKa = 3.39IAYY349 pKa = 9.74GYY351 pKa = 10.47AQLADD356 pKa = 3.43TDD358 pKa = 4.25RR359 pKa = 11.84NAEE362 pKa = 4.12AFLKK366 pKa = 10.95SNSQAKK372 pKa = 8.57YY373 pKa = 10.44VKK375 pKa = 10.13DD376 pKa = 3.53CGVMVRR382 pKa = 11.84LYY384 pKa = 10.73KK385 pKa = 10.13KK386 pKa = 10.81AEE388 pKa = 3.91MQRR391 pKa = 11.84TSMKK395 pKa = 10.27DD396 pKa = 2.91WIKK399 pKa = 10.62RR400 pKa = 11.84RR401 pKa = 11.84CEE403 pKa = 3.93EE404 pKa = 4.07VDD406 pKa = 3.84GDD408 pKa = 4.32GDD410 pKa = 3.39WRR412 pKa = 11.84PIVSFLKK419 pKa = 10.72FQGIDD424 pKa = 3.06MAVFISVLKK433 pKa = 10.77NFLKK437 pKa = 10.92GIPKK441 pKa = 9.61QNCIVICGPPNTGKK455 pKa = 10.6SLFSVSLIQFLSGKK469 pKa = 10.0VISFVNSKK477 pKa = 8.57SHH479 pKa = 7.02FWLQPLGEE487 pKa = 4.64CKK489 pKa = 10.17VALLDD494 pKa = 5.24DD495 pKa = 4.1ATGPAWDD502 pKa = 4.22YY503 pKa = 11.58FDD505 pKa = 3.62TFLRR509 pKa = 11.84NLVDD513 pKa = 4.08GNPISLDD520 pKa = 3.89CKK522 pKa = 10.46HH523 pKa = 6.62KK524 pKa = 11.09APTQVKK530 pKa = 9.21CPPLIITSNIDD541 pKa = 2.95VTASDD546 pKa = 3.02RR547 pKa = 11.84WKK549 pKa = 10.49YY550 pKa = 10.0LHH552 pKa = 7.01SRR554 pKa = 11.84LKK556 pKa = 11.04LFTFSNPCPLTEE568 pKa = 4.04QGEE571 pKa = 4.37PAFNLTPVNWKK582 pKa = 10.37CFFKK586 pKa = 10.76RR587 pKa = 11.84LKK589 pKa = 9.68TRR591 pKa = 11.84LGLADD596 pKa = 5.7DD597 pKa = 4.35EE598 pKa = 5.48GEE600 pKa = 4.18DD601 pKa = 4.1HH602 pKa = 7.37GEE604 pKa = 3.89SQQPLKK610 pKa = 10.17CTARR614 pKa = 11.84GTDD617 pKa = 3.33GFVV620 pKa = 2.77
Molecular weight: 68.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2VIS3|F2VIS3_9PAPI E4 protein (Fragment) OS=Phocoena phocoena papillomavirus 4 OX=706527 GN=E4 PE=4 SV=1
LL1 pKa = 6.64TLNLLLAQPIPRR13 pKa = 11.84PPRR16 pKa = 11.84VPVPRR21 pKa = 11.84SPYY24 pKa = 10.5LDD26 pKa = 3.72LLGHH30 pKa = 6.9IPRR33 pKa = 11.84TPPQQHH39 pKa = 6.67APPTCPRR46 pKa = 11.84LQRR49 pKa = 11.84RR50 pKa = 11.84GNPGRR55 pKa = 11.84QPLPRR60 pKa = 11.84ADD62 pKa = 3.81KK63 pKa = 11.0DD64 pKa = 4.06PIRR67 pKa = 11.84KK68 pKa = 9.29RR69 pKa = 11.84LWDD72 pKa = 3.77SDD74 pKa = 3.6DD75 pKa = 4.28SEE77 pKa = 4.57NKK79 pKa = 10.17EE80 pKa = 4.47NIPPLQQQGPYY91 pKa = 9.22TVDD94 pKa = 4.36FPPIGPLDD102 pKa = 3.9PPVTVTLTVTFQQ114 pKa = 2.95
LL1 pKa = 6.64TLNLLLAQPIPRR13 pKa = 11.84PPRR16 pKa = 11.84VPVPRR21 pKa = 11.84SPYY24 pKa = 10.5LDD26 pKa = 3.72LLGHH30 pKa = 6.9IPRR33 pKa = 11.84TPPQQHH39 pKa = 6.67APPTCPRR46 pKa = 11.84LQRR49 pKa = 11.84RR50 pKa = 11.84GNPGRR55 pKa = 11.84QPLPRR60 pKa = 11.84ADD62 pKa = 3.81KK63 pKa = 11.0DD64 pKa = 4.06PIRR67 pKa = 11.84KK68 pKa = 9.29RR69 pKa = 11.84LWDD72 pKa = 3.77SDD74 pKa = 3.6DD75 pKa = 4.28SEE77 pKa = 4.57NKK79 pKa = 10.17EE80 pKa = 4.47NIPPLQQQGPYY91 pKa = 9.22TVDD94 pKa = 4.36FPPIGPLDD102 pKa = 3.9PPVTVTLTVTFQQ114 pKa = 2.95
Molecular weight: 12.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2281 |
114 |
620 |
380.2 |
42.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.612 ± 0.666 | 2.499 ± 0.604 |
6.357 ± 0.644 | 5.042 ± 0.414 |
3.902 ± 0.455 | 7.541 ± 0.841 |
1.929 ± 0.288 | 4.603 ± 0.551 |
5.48 ± 0.621 | 8.242 ± 0.664 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.666 ± 0.285 | 3.683 ± 0.287 |
7.321 ± 1.289 | 4.428 ± 0.382 |
5.305 ± 0.602 | 7.453 ± 0.667 |
7.716 ± 0.752 | 6.401 ± 0.235 |
1.622 ± 0.339 | 3.2 ± 0.58 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
