
Rhodococcus coprophilus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Nocardiaceae; Rhodococcus
Average proteome isoelectric point is 5.85
Get precalculated fractions of proteins
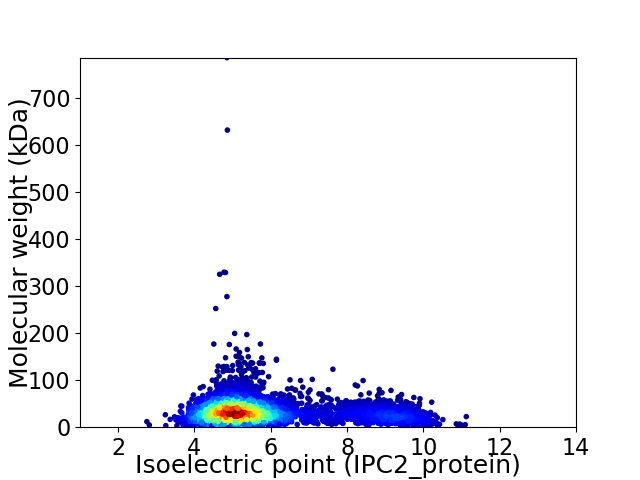
Virtual 2D-PAGE plot for 4163 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2X4WRT6|A0A2X4WRT6_9NOCA Beta lactamase OS=Rhodococcus coprophilus OX=38310 GN=NCTC10994_01188 PE=4 SV=1
MM1 pKa = 7.51KK2 pKa = 9.81KK3 pKa = 10.05LHH5 pKa = 6.7IGAIAAVAALTVSACGSSGGDD26 pKa = 3.14ASGDD30 pKa = 3.32AAAPTGAPIVIGAQAPIKK48 pKa = 10.49GATAYY53 pKa = 8.43PQTAYY58 pKa = 10.56GLEE61 pKa = 4.29AGVHH65 pKa = 4.84YY66 pKa = 10.9VNEE69 pKa = 4.11VLGGINGRR77 pKa = 11.84PLEE80 pKa = 3.99VDD82 pKa = 3.3VCGGDD87 pKa = 3.69GSPEE91 pKa = 3.89TSINCANGFVSKK103 pKa = 10.6NVPLVIDD110 pKa = 4.71AVDD113 pKa = 3.46QSMGAAVPILGSAGIPIIGTLAGSFVADD141 pKa = 3.29AAEE144 pKa = 4.19YY145 pKa = 9.95GQAFYY150 pKa = 8.59FTGPVSFAALASMSALDD167 pKa = 3.86TMGKK171 pKa = 6.38TTASLAVNEE180 pKa = 4.97SPTSHH185 pKa = 7.02AYY187 pKa = 10.27VDD189 pKa = 4.02DD190 pKa = 5.75LIMPIADD197 pKa = 3.23GMGIDD202 pKa = 4.65VSVQYY207 pKa = 10.47PPASGANLNVVAATQLADD225 pKa = 3.76NPDD228 pKa = 3.56MAGVIALPEE237 pKa = 4.75DD238 pKa = 4.03GCTSLFQALRR248 pKa = 11.84RR249 pKa = 11.84QGYY252 pKa = 9.65EE253 pKa = 3.72DD254 pKa = 4.35TILAGSCSQFIDD266 pKa = 4.49EE267 pKa = 5.18MGTDD271 pKa = 3.31AAGIVTQPRR280 pKa = 11.84LWVPLAKK287 pKa = 10.2EE288 pKa = 3.91SAPAEE293 pKa = 4.19VQEE296 pKa = 4.23QLDD299 pKa = 3.84IFADD303 pKa = 3.44AMDD306 pKa = 3.61AVGYY310 pKa = 10.57GNEE313 pKa = 3.89LSSRR317 pKa = 11.84SIYY320 pKa = 10.13TFAAIVNIAQILSDD334 pKa = 3.73HH335 pKa = 6.53GGEE338 pKa = 3.91ITAGTVTDD346 pKa = 3.73AFKK349 pKa = 10.72AVKK352 pKa = 10.05DD353 pKa = 4.49FPSFAGPTVTCDD365 pKa = 3.08GQQWPGVATTCNAQAIFFEE384 pKa = 4.41VQEE387 pKa = 4.83DD388 pKa = 4.1GTLRR392 pKa = 11.84SVDD395 pKa = 3.38EE396 pKa = 4.49NGYY399 pKa = 10.1IDD401 pKa = 5.92LDD403 pKa = 4.03PSLMPASS410 pKa = 3.79
MM1 pKa = 7.51KK2 pKa = 9.81KK3 pKa = 10.05LHH5 pKa = 6.7IGAIAAVAALTVSACGSSGGDD26 pKa = 3.14ASGDD30 pKa = 3.32AAAPTGAPIVIGAQAPIKK48 pKa = 10.49GATAYY53 pKa = 8.43PQTAYY58 pKa = 10.56GLEE61 pKa = 4.29AGVHH65 pKa = 4.84YY66 pKa = 10.9VNEE69 pKa = 4.11VLGGINGRR77 pKa = 11.84PLEE80 pKa = 3.99VDD82 pKa = 3.3VCGGDD87 pKa = 3.69GSPEE91 pKa = 3.89TSINCANGFVSKK103 pKa = 10.6NVPLVIDD110 pKa = 4.71AVDD113 pKa = 3.46QSMGAAVPILGSAGIPIIGTLAGSFVADD141 pKa = 3.29AAEE144 pKa = 4.19YY145 pKa = 9.95GQAFYY150 pKa = 8.59FTGPVSFAALASMSALDD167 pKa = 3.86TMGKK171 pKa = 6.38TTASLAVNEE180 pKa = 4.97SPTSHH185 pKa = 7.02AYY187 pKa = 10.27VDD189 pKa = 4.02DD190 pKa = 5.75LIMPIADD197 pKa = 3.23GMGIDD202 pKa = 4.65VSVQYY207 pKa = 10.47PPASGANLNVVAATQLADD225 pKa = 3.76NPDD228 pKa = 3.56MAGVIALPEE237 pKa = 4.75DD238 pKa = 4.03GCTSLFQALRR248 pKa = 11.84RR249 pKa = 11.84QGYY252 pKa = 9.65EE253 pKa = 3.72DD254 pKa = 4.35TILAGSCSQFIDD266 pKa = 4.49EE267 pKa = 5.18MGTDD271 pKa = 3.31AAGIVTQPRR280 pKa = 11.84LWVPLAKK287 pKa = 10.2EE288 pKa = 3.91SAPAEE293 pKa = 4.19VQEE296 pKa = 4.23QLDD299 pKa = 3.84IFADD303 pKa = 3.44AMDD306 pKa = 3.61AVGYY310 pKa = 10.57GNEE313 pKa = 3.89LSSRR317 pKa = 11.84SIYY320 pKa = 10.13TFAAIVNIAQILSDD334 pKa = 3.73HH335 pKa = 6.53GGEE338 pKa = 3.91ITAGTVTDD346 pKa = 3.73AFKK349 pKa = 10.72AVKK352 pKa = 10.05DD353 pKa = 4.49FPSFAGPTVTCDD365 pKa = 3.08GQQWPGVATTCNAQAIFFEE384 pKa = 4.41VQEE387 pKa = 4.83DD388 pKa = 4.1GTLRR392 pKa = 11.84SVDD395 pKa = 3.38EE396 pKa = 4.49NGYY399 pKa = 10.1IDD401 pKa = 5.92LDD403 pKa = 4.03PSLMPASS410 pKa = 3.79
Molecular weight: 41.71 kDa
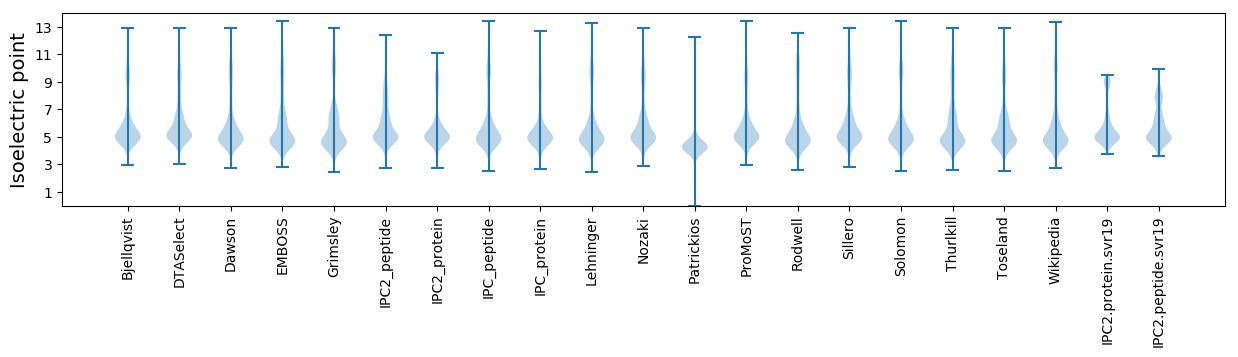
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2X4TLZ1|A0A2X4TLZ1_9NOCA Uncharacterized protein OS=Rhodococcus coprophilus OX=38310 GN=NCTC10994_00316 PE=4 SV=1
MM1 pKa = 7.47AHH3 pKa = 5.88RR4 pKa = 11.84TAIEE8 pKa = 4.21VIPAARR14 pKa = 11.84RR15 pKa = 11.84IGILALRR22 pKa = 11.84APTIGRR28 pKa = 11.84TLVPIVAPTPAPTAVPKK45 pKa = 10.32VVRR48 pKa = 11.84ISGRR52 pKa = 11.84IVVRR56 pKa = 11.84RR57 pKa = 11.84VVLIVVPRR65 pKa = 11.84VVRR68 pKa = 11.84ISVPIVVPRR77 pKa = 11.84VVLIAAPRR85 pKa = 11.84VVRR88 pKa = 11.84ISVPIVVPRR97 pKa = 11.84AVLIVVPKK105 pKa = 9.86VVRR108 pKa = 11.84TTVPIVVRR116 pKa = 11.84RR117 pKa = 11.84VVRR120 pKa = 11.84ISGRR124 pKa = 11.84IVVRR128 pKa = 11.84KK129 pKa = 9.21AVPIVVRR136 pKa = 11.84RR137 pKa = 11.84VVRR140 pKa = 11.84ITAPIVVPKK149 pKa = 10.39AVPNTGTTPVPIVVRR164 pKa = 11.84TLVPIAGLRR173 pKa = 11.84DD174 pKa = 3.33VRR176 pKa = 11.84TLVPIAAPTLARR188 pKa = 11.84IDD190 pKa = 3.56GQKK193 pKa = 9.8PRR195 pKa = 11.84RR196 pKa = 11.84RR197 pKa = 11.84TPGRR201 pKa = 11.84SRR203 pKa = 11.84TARR206 pKa = 11.84APRR209 pKa = 3.72
MM1 pKa = 7.47AHH3 pKa = 5.88RR4 pKa = 11.84TAIEE8 pKa = 4.21VIPAARR14 pKa = 11.84RR15 pKa = 11.84IGILALRR22 pKa = 11.84APTIGRR28 pKa = 11.84TLVPIVAPTPAPTAVPKK45 pKa = 10.32VVRR48 pKa = 11.84ISGRR52 pKa = 11.84IVVRR56 pKa = 11.84RR57 pKa = 11.84VVLIVVPRR65 pKa = 11.84VVRR68 pKa = 11.84ISVPIVVPRR77 pKa = 11.84VVLIAAPRR85 pKa = 11.84VVRR88 pKa = 11.84ISVPIVVPRR97 pKa = 11.84AVLIVVPKK105 pKa = 9.86VVRR108 pKa = 11.84TTVPIVVRR116 pKa = 11.84RR117 pKa = 11.84VVRR120 pKa = 11.84ISGRR124 pKa = 11.84IVVRR128 pKa = 11.84KK129 pKa = 9.21AVPIVVRR136 pKa = 11.84RR137 pKa = 11.84VVRR140 pKa = 11.84ITAPIVVPKK149 pKa = 10.39AVPNTGTTPVPIVVRR164 pKa = 11.84TLVPIAGLRR173 pKa = 11.84DD174 pKa = 3.33VRR176 pKa = 11.84TLVPIAAPTLARR188 pKa = 11.84IDD190 pKa = 3.56GQKK193 pKa = 9.8PRR195 pKa = 11.84RR196 pKa = 11.84RR197 pKa = 11.84TPGRR201 pKa = 11.84SRR203 pKa = 11.84TARR206 pKa = 11.84APRR209 pKa = 3.72
Molecular weight: 22.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1371707 |
29 |
7364 |
329.5 |
35.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.649 ± 0.052 | 0.739 ± 0.011 |
6.52 ± 0.033 | 5.813 ± 0.036 |
3.021 ± 0.02 | 9.002 ± 0.035 |
2.15 ± 0.02 | 4.032 ± 0.028 |
2.032 ± 0.028 | 9.961 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.911 ± 0.014 | 1.972 ± 0.02 |
5.706 ± 0.031 | 2.6 ± 0.023 |
7.552 ± 0.041 | 5.53 ± 0.022 |
6.195 ± 0.03 | 9.16 ± 0.037 |
1.412 ± 0.013 | 2.04 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
