
Roseburia sp. CAG:100
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Roseburia; environmental samples
Average proteome isoelectric point is 5.86
Get precalculated fractions of proteins
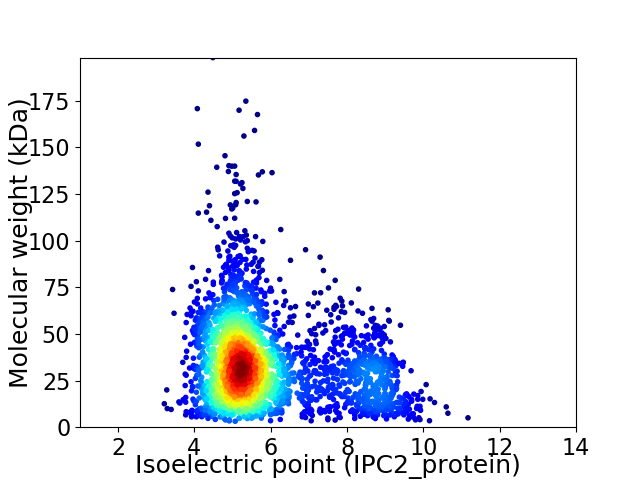
Virtual 2D-PAGE plot for 2901 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
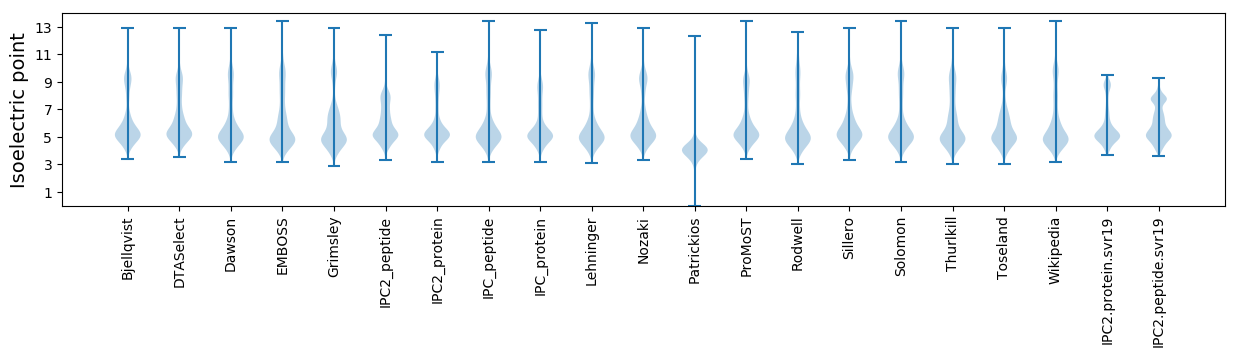
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7R103|R7R103_9FIRM Pseudouridine synthase OS=Roseburia sp. CAG:100 OX=1262940 GN=BN450_01026 PE=3 SV=1
MM1 pKa = 7.8LEE3 pKa = 4.3TPVEE7 pKa = 4.22SLDD10 pKa = 3.63PQQATDD16 pKa = 3.57GTSFEE21 pKa = 4.7VIADD25 pKa = 3.82YY26 pKa = 11.2TDD28 pKa = 4.32GLMQMDD34 pKa = 4.94ADD36 pKa = 4.24GQAVPALAEE45 pKa = 4.41SYY47 pKa = 10.85DD48 pKa = 3.9LSDD51 pKa = 5.66DD52 pKa = 3.51GLTYY56 pKa = 10.34TFHH59 pKa = 8.08IRR61 pKa = 11.84DD62 pKa = 3.68DD63 pKa = 4.61ANWSNGTPVTAADD76 pKa = 5.8FVFAWQRR83 pKa = 11.84AADD86 pKa = 3.83PEE88 pKa = 4.46VASEE92 pKa = 3.86YY93 pKa = 10.9SYY95 pKa = 10.83MLSDD99 pKa = 3.75IGQIKK104 pKa = 9.57NAADD108 pKa = 3.98IIAGNKK114 pKa = 9.4DD115 pKa = 3.11KK116 pKa = 11.76SEE118 pKa = 4.54LGVTAVDD125 pKa = 4.28DD126 pKa = 3.6KK127 pKa = 11.27TLQVEE132 pKa = 4.37LNVPVSYY139 pKa = 10.22FLSLMYY145 pKa = 10.48FPTFYY150 pKa = 10.39PVNEE154 pKa = 4.34EE155 pKa = 4.29FFTSCGDD162 pKa = 3.56TFATSPEE169 pKa = 4.25TVLSNGAFVLDD180 pKa = 4.22SYY182 pKa = 11.36EE183 pKa = 3.99PAATAFHH190 pKa = 6.21LTKK193 pKa = 10.88NADD196 pKa = 3.57YY197 pKa = 11.02YY198 pKa = 11.4NADD201 pKa = 4.01SIQLAGLNYY210 pKa = 10.12QVIQDD215 pKa = 3.85SQQALMSYY223 pKa = 7.98QTGALDD229 pKa = 3.51TTLVNGEE236 pKa = 4.11QVDD239 pKa = 4.0QVKK242 pKa = 10.7DD243 pKa = 3.69DD244 pKa = 4.81PEE246 pKa = 4.16FQAIGAGYY254 pKa = 9.63LWYY257 pKa = 10.61VSPNIKK263 pKa = 10.04NVPEE267 pKa = 3.95LANLNIRR274 pKa = 11.84MALTMAINRR283 pKa = 11.84EE284 pKa = 4.26AITTDD289 pKa = 3.18VLKK292 pKa = 11.03DD293 pKa = 3.64GSTPTYY299 pKa = 8.31TAVPIEE305 pKa = 4.46FAAGPDD311 pKa = 3.62GSDD314 pKa = 3.67FSGDD318 pKa = 3.12QSKK321 pKa = 10.65FSDD324 pKa = 3.62VCAFDD329 pKa = 3.91ADD331 pKa = 3.7KK332 pKa = 11.09AAEE335 pKa = 3.98YY336 pKa = 8.67WQAGLDD342 pKa = 3.7EE343 pKa = 5.42LGVTEE348 pKa = 5.1LALDD352 pKa = 3.84MVVDD356 pKa = 5.47ADD358 pKa = 4.76DD359 pKa = 5.1APQKK363 pKa = 9.76VAQVLKK369 pKa = 9.23EE370 pKa = 3.9QWEE373 pKa = 4.5TTLPGLTVTLTIEE386 pKa = 4.16PKK388 pKa = 9.56KK389 pKa = 10.88QRR391 pKa = 11.84VQDD394 pKa = 3.73MQDD397 pKa = 2.95GNFQLGLTRR406 pKa = 11.84WGPDD410 pKa = 3.01YY411 pKa = 10.98ADD413 pKa = 3.44PMTYY417 pKa = 10.55LGMWVTDD424 pKa = 3.28NSNNYY429 pKa = 8.94GFWSNAEE436 pKa = 3.52YY437 pKa = 10.72DD438 pKa = 4.24AIIDD442 pKa = 3.72EE443 pKa = 4.83CTTGDD448 pKa = 3.77LCTDD452 pKa = 3.57AEE454 pKa = 4.59GRR456 pKa = 11.84WARR459 pKa = 11.84LYY461 pKa = 10.54DD462 pKa = 3.57AEE464 pKa = 5.01KK465 pKa = 10.45IVMDD469 pKa = 4.04EE470 pKa = 3.89AVIFPLYY477 pKa = 7.05TQCNAEE483 pKa = 4.11MMSSKK488 pKa = 9.18VTGVEE493 pKa = 4.08FHH495 pKa = 6.8PVALNRR501 pKa = 11.84VYY503 pKa = 11.1KK504 pKa = 10.67NAVKK508 pKa = 10.85AEE510 pKa = 3.93
MM1 pKa = 7.8LEE3 pKa = 4.3TPVEE7 pKa = 4.22SLDD10 pKa = 3.63PQQATDD16 pKa = 3.57GTSFEE21 pKa = 4.7VIADD25 pKa = 3.82YY26 pKa = 11.2TDD28 pKa = 4.32GLMQMDD34 pKa = 4.94ADD36 pKa = 4.24GQAVPALAEE45 pKa = 4.41SYY47 pKa = 10.85DD48 pKa = 3.9LSDD51 pKa = 5.66DD52 pKa = 3.51GLTYY56 pKa = 10.34TFHH59 pKa = 8.08IRR61 pKa = 11.84DD62 pKa = 3.68DD63 pKa = 4.61ANWSNGTPVTAADD76 pKa = 5.8FVFAWQRR83 pKa = 11.84AADD86 pKa = 3.83PEE88 pKa = 4.46VASEE92 pKa = 3.86YY93 pKa = 10.9SYY95 pKa = 10.83MLSDD99 pKa = 3.75IGQIKK104 pKa = 9.57NAADD108 pKa = 3.98IIAGNKK114 pKa = 9.4DD115 pKa = 3.11KK116 pKa = 11.76SEE118 pKa = 4.54LGVTAVDD125 pKa = 4.28DD126 pKa = 3.6KK127 pKa = 11.27TLQVEE132 pKa = 4.37LNVPVSYY139 pKa = 10.22FLSLMYY145 pKa = 10.48FPTFYY150 pKa = 10.39PVNEE154 pKa = 4.34EE155 pKa = 4.29FFTSCGDD162 pKa = 3.56TFATSPEE169 pKa = 4.25TVLSNGAFVLDD180 pKa = 4.22SYY182 pKa = 11.36EE183 pKa = 3.99PAATAFHH190 pKa = 6.21LTKK193 pKa = 10.88NADD196 pKa = 3.57YY197 pKa = 11.02YY198 pKa = 11.4NADD201 pKa = 4.01SIQLAGLNYY210 pKa = 10.12QVIQDD215 pKa = 3.85SQQALMSYY223 pKa = 7.98QTGALDD229 pKa = 3.51TTLVNGEE236 pKa = 4.11QVDD239 pKa = 4.0QVKK242 pKa = 10.7DD243 pKa = 3.69DD244 pKa = 4.81PEE246 pKa = 4.16FQAIGAGYY254 pKa = 9.63LWYY257 pKa = 10.61VSPNIKK263 pKa = 10.04NVPEE267 pKa = 3.95LANLNIRR274 pKa = 11.84MALTMAINRR283 pKa = 11.84EE284 pKa = 4.26AITTDD289 pKa = 3.18VLKK292 pKa = 11.03DD293 pKa = 3.64GSTPTYY299 pKa = 8.31TAVPIEE305 pKa = 4.46FAAGPDD311 pKa = 3.62GSDD314 pKa = 3.67FSGDD318 pKa = 3.12QSKK321 pKa = 10.65FSDD324 pKa = 3.62VCAFDD329 pKa = 3.91ADD331 pKa = 3.7KK332 pKa = 11.09AAEE335 pKa = 3.98YY336 pKa = 8.67WQAGLDD342 pKa = 3.7EE343 pKa = 5.42LGVTEE348 pKa = 5.1LALDD352 pKa = 3.84MVVDD356 pKa = 5.47ADD358 pKa = 4.76DD359 pKa = 5.1APQKK363 pKa = 9.76VAQVLKK369 pKa = 9.23EE370 pKa = 3.9QWEE373 pKa = 4.5TTLPGLTVTLTIEE386 pKa = 4.16PKK388 pKa = 9.56KK389 pKa = 10.88QRR391 pKa = 11.84VQDD394 pKa = 3.73MQDD397 pKa = 2.95GNFQLGLTRR406 pKa = 11.84WGPDD410 pKa = 3.01YY411 pKa = 10.98ADD413 pKa = 3.44PMTYY417 pKa = 10.55LGMWVTDD424 pKa = 3.28NSNNYY429 pKa = 8.94GFWSNAEE436 pKa = 3.52YY437 pKa = 10.72DD438 pKa = 4.24AIIDD442 pKa = 3.72EE443 pKa = 4.83CTTGDD448 pKa = 3.77LCTDD452 pKa = 3.57AEE454 pKa = 4.59GRR456 pKa = 11.84WARR459 pKa = 11.84LYY461 pKa = 10.54DD462 pKa = 3.57AEE464 pKa = 5.01KK465 pKa = 10.45IVMDD469 pKa = 4.04EE470 pKa = 3.89AVIFPLYY477 pKa = 7.05TQCNAEE483 pKa = 4.11MMSSKK488 pKa = 9.18VTGVEE493 pKa = 4.08FHH495 pKa = 6.8PVALNRR501 pKa = 11.84VYY503 pKa = 11.1KK504 pKa = 10.67NAVKK508 pKa = 10.85AEE510 pKa = 3.93
Molecular weight: 56.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7RA20|R7RA20_9FIRM ABC transmembrane type-1 domain-containing protein OS=Roseburia sp. CAG:100 OX=1262940 GN=BN450_00576 PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.71MTFQPKK8 pKa = 7.43TRR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.26VHH16 pKa = 5.59GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.67KK2 pKa = 8.71MTFQPKK8 pKa = 7.43TRR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.26VHH16 pKa = 5.59GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
925423 |
29 |
1784 |
319.0 |
35.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.521 ± 0.051 | 1.482 ± 0.017 |
5.842 ± 0.035 | 7.522 ± 0.039 |
3.937 ± 0.029 | 6.708 ± 0.035 |
1.848 ± 0.023 | 7.596 ± 0.037 |
6.514 ± 0.039 | 8.704 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.285 ± 0.023 | 4.523 ± 0.029 |
3.11 ± 0.025 | 3.741 ± 0.032 |
4.212 ± 0.037 | 5.588 ± 0.037 |
5.536 ± 0.032 | 6.828 ± 0.033 |
0.957 ± 0.014 | 4.541 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
