
Equus caballus papillomavirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyoiotapapillomavirus; Dyoiotapapillomavirus 1
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
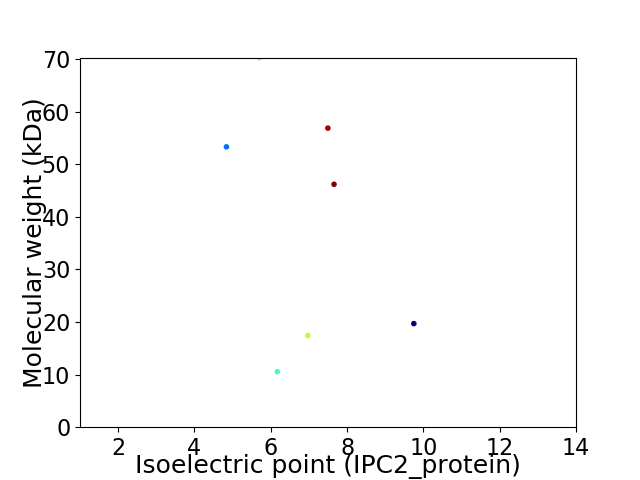
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E7C0H6|E7C0H6_9PAPI Major capsid protein L1 OS=Equus caballus papillomavirus 3 OX=940834 GN=L1 PE=3 SV=1
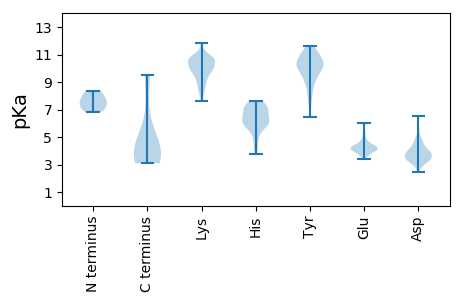
MM1 pKa = 7.1TSGRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.55RR8 pKa = 11.84RR9 pKa = 11.84AADD12 pKa = 3.51TTDD15 pKa = 3.03TPPRR19 pKa = 11.84KK20 pKa = 9.3RR21 pKa = 11.84RR22 pKa = 11.84PRR24 pKa = 11.84AAVEE28 pKa = 4.11DD29 pKa = 4.33IYY31 pKa = 11.17RR32 pKa = 11.84GCKK35 pKa = 9.21PFNTCPEE42 pKa = 4.13DD43 pKa = 3.46VVNRR47 pKa = 11.84VEE49 pKa = 4.1NKK51 pKa = 7.59TWADD55 pKa = 4.73RR56 pKa = 11.84LLQWLGSVIYY66 pKa = 10.66LGGLGIGSGRR76 pKa = 11.84GTGGGTGYY84 pKa = 10.72RR85 pKa = 11.84PLGSTNRR92 pKa = 11.84GVVVGGGSRR101 pKa = 11.84AVRR104 pKa = 11.84PPPLVVDD111 pKa = 4.33TIGPVDD117 pKa = 3.81AEE119 pKa = 4.29VLEE122 pKa = 4.72MIPLQPLEE130 pKa = 4.37PGGPSVVSGSDD141 pKa = 2.97GGVLVSEE148 pKa = 5.19GGSLPPEE155 pKa = 4.07IPGVLDD161 pKa = 3.42GGGPEE166 pKa = 4.3DD167 pKa = 5.1SIPARR172 pKa = 11.84DD173 pKa = 3.62PAVLQVGEE181 pKa = 4.24GDD183 pKa = 3.44TGNSHH188 pKa = 7.35VITPAIEE195 pKa = 3.92RR196 pKa = 11.84PGFSGSSRR204 pKa = 11.84DD205 pKa = 2.87SFYY208 pKa = 10.78IVDD211 pKa = 3.61SGSRR215 pKa = 11.84GSVVGEE221 pKa = 4.38DD222 pKa = 3.52IEE224 pKa = 5.82LVDD227 pKa = 5.48LPRR230 pKa = 11.84ASTPEE235 pKa = 3.99TTGDD239 pKa = 3.34ALTRR243 pKa = 11.84GLGSRR248 pKa = 11.84KK249 pKa = 8.24YY250 pKa = 8.36QQVYY254 pKa = 9.29VEE256 pKa = 4.91NPAFVYY262 pKa = 10.81NPTEE266 pKa = 3.8LVAFGDD272 pKa = 3.87TWDD275 pKa = 3.92GATDD279 pKa = 3.57SFSYY283 pKa = 10.88DD284 pKa = 3.41PVVASPQAAPDD295 pKa = 3.64EE296 pKa = 4.54LFTDD300 pKa = 4.11IVHH303 pKa = 7.51LGRR306 pKa = 11.84QMYY309 pKa = 9.68EE310 pKa = 3.69RR311 pKa = 11.84GRR313 pKa = 11.84EE314 pKa = 3.74GLLRR318 pKa = 11.84VGRR321 pKa = 11.84VGRR324 pKa = 11.84RR325 pKa = 11.84GTIQTRR331 pKa = 11.84AGTQIGPQVHH341 pKa = 6.66FFHH344 pKa = 7.51DD345 pKa = 4.74LSPILPVQEE354 pKa = 4.28EE355 pKa = 4.81VEE357 pKa = 4.32LTTFPRR363 pKa = 11.84APEE366 pKa = 3.98FEE368 pKa = 4.45STNSEE373 pKa = 3.57EE374 pKa = 4.31TAFTEE379 pKa = 4.55VDD381 pKa = 3.82LQSEE385 pKa = 4.33PSSYY389 pKa = 11.13SDD391 pKa = 3.22TYY393 pKa = 11.22LVEE396 pKa = 5.22DD397 pKa = 4.83DD398 pKa = 4.17SVSVTGHH405 pKa = 6.54LVFSDD410 pKa = 3.48EE411 pKa = 4.83GGSLTDD417 pKa = 3.41NVHH420 pKa = 7.22PINVPLRR427 pKa = 11.84GSAGVGLLSVQGTTDD442 pKa = 3.26NLTSNSLTPALQPPQPGGGDD462 pKa = 3.19RR463 pKa = 11.84EE464 pKa = 4.64VVVNMYY470 pKa = 9.42PYY472 pKa = 10.79SIMSFLHH479 pKa = 4.2YY480 pKa = 9.76RR481 pKa = 11.84RR482 pKa = 11.84RR483 pKa = 11.84RR484 pKa = 11.84KK485 pKa = 9.5RR486 pKa = 11.84GYY488 pKa = 10.24VYY490 pKa = 10.6FSDD493 pKa = 4.34VILAII498 pKa = 4.24
MM1 pKa = 7.1TSGRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.55RR8 pKa = 11.84RR9 pKa = 11.84AADD12 pKa = 3.51TTDD15 pKa = 3.03TPPRR19 pKa = 11.84KK20 pKa = 9.3RR21 pKa = 11.84RR22 pKa = 11.84PRR24 pKa = 11.84AAVEE28 pKa = 4.11DD29 pKa = 4.33IYY31 pKa = 11.17RR32 pKa = 11.84GCKK35 pKa = 9.21PFNTCPEE42 pKa = 4.13DD43 pKa = 3.46VVNRR47 pKa = 11.84VEE49 pKa = 4.1NKK51 pKa = 7.59TWADD55 pKa = 4.73RR56 pKa = 11.84LLQWLGSVIYY66 pKa = 10.66LGGLGIGSGRR76 pKa = 11.84GTGGGTGYY84 pKa = 10.72RR85 pKa = 11.84PLGSTNRR92 pKa = 11.84GVVVGGGSRR101 pKa = 11.84AVRR104 pKa = 11.84PPPLVVDD111 pKa = 4.33TIGPVDD117 pKa = 3.81AEE119 pKa = 4.29VLEE122 pKa = 4.72MIPLQPLEE130 pKa = 4.37PGGPSVVSGSDD141 pKa = 2.97GGVLVSEE148 pKa = 5.19GGSLPPEE155 pKa = 4.07IPGVLDD161 pKa = 3.42GGGPEE166 pKa = 4.3DD167 pKa = 5.1SIPARR172 pKa = 11.84DD173 pKa = 3.62PAVLQVGEE181 pKa = 4.24GDD183 pKa = 3.44TGNSHH188 pKa = 7.35VITPAIEE195 pKa = 3.92RR196 pKa = 11.84PGFSGSSRR204 pKa = 11.84DD205 pKa = 2.87SFYY208 pKa = 10.78IVDD211 pKa = 3.61SGSRR215 pKa = 11.84GSVVGEE221 pKa = 4.38DD222 pKa = 3.52IEE224 pKa = 5.82LVDD227 pKa = 5.48LPRR230 pKa = 11.84ASTPEE235 pKa = 3.99TTGDD239 pKa = 3.34ALTRR243 pKa = 11.84GLGSRR248 pKa = 11.84KK249 pKa = 8.24YY250 pKa = 8.36QQVYY254 pKa = 9.29VEE256 pKa = 4.91NPAFVYY262 pKa = 10.81NPTEE266 pKa = 3.8LVAFGDD272 pKa = 3.87TWDD275 pKa = 3.92GATDD279 pKa = 3.57SFSYY283 pKa = 10.88DD284 pKa = 3.41PVVASPQAAPDD295 pKa = 3.64EE296 pKa = 4.54LFTDD300 pKa = 4.11IVHH303 pKa = 7.51LGRR306 pKa = 11.84QMYY309 pKa = 9.68EE310 pKa = 3.69RR311 pKa = 11.84GRR313 pKa = 11.84EE314 pKa = 3.74GLLRR318 pKa = 11.84VGRR321 pKa = 11.84VGRR324 pKa = 11.84RR325 pKa = 11.84GTIQTRR331 pKa = 11.84AGTQIGPQVHH341 pKa = 6.66FFHH344 pKa = 7.51DD345 pKa = 4.74LSPILPVQEE354 pKa = 4.28EE355 pKa = 4.81VEE357 pKa = 4.32LTTFPRR363 pKa = 11.84APEE366 pKa = 3.98FEE368 pKa = 4.45STNSEE373 pKa = 3.57EE374 pKa = 4.31TAFTEE379 pKa = 4.55VDD381 pKa = 3.82LQSEE385 pKa = 4.33PSSYY389 pKa = 11.13SDD391 pKa = 3.22TYY393 pKa = 11.22LVEE396 pKa = 5.22DD397 pKa = 4.83DD398 pKa = 4.17SVSVTGHH405 pKa = 6.54LVFSDD410 pKa = 3.48EE411 pKa = 4.83GGSLTDD417 pKa = 3.41NVHH420 pKa = 7.22PINVPLRR427 pKa = 11.84GSAGVGLLSVQGTTDD442 pKa = 3.26NLTSNSLTPALQPPQPGGGDD462 pKa = 3.19RR463 pKa = 11.84EE464 pKa = 4.64VVVNMYY470 pKa = 9.42PYY472 pKa = 10.79SIMSFLHH479 pKa = 4.2YY480 pKa = 9.76RR481 pKa = 11.84RR482 pKa = 11.84RR483 pKa = 11.84RR484 pKa = 11.84KK485 pKa = 9.5RR486 pKa = 11.84GYY488 pKa = 10.24VYY490 pKa = 10.6FSDD493 pKa = 4.34VILAII498 pKa = 4.24
Molecular weight: 53.31 kDa
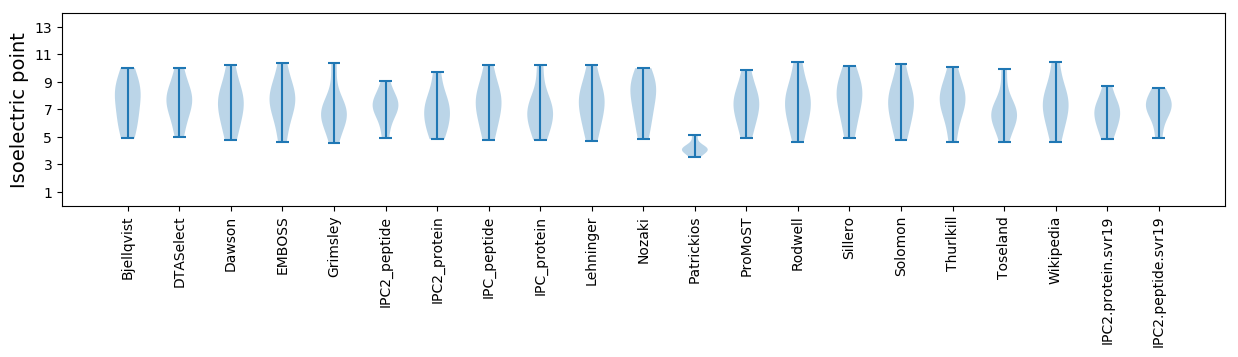
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E7C0H5|E7C0H5_9PAPI Minor capsid protein L2 OS=Equus caballus papillomavirus 3 OX=940834 GN=L2 PE=3 SV=1
II1 pKa = 7.28PKK3 pKa = 10.15GYY5 pKa = 8.95TIVRR9 pKa = 11.84GPLKK13 pKa = 10.66SIMRR17 pKa = 11.84TLQLKK22 pKa = 9.72QKK24 pKa = 9.13STLKK28 pKa = 10.69QMFGKK33 pKa = 10.1SCFKK37 pKa = 10.22TKK39 pKa = 10.5HH40 pKa = 5.74FLLSPAEE47 pKa = 4.08LPRR50 pKa = 11.84RR51 pKa = 11.84TPQSLLDD58 pKa = 3.93LAYY61 pKa = 10.36QAATSRR67 pKa = 11.84TAQQRR72 pKa = 11.84SPVYY76 pKa = 10.08FPLRR80 pKa = 11.84LPPCLLNSSLYY91 pKa = 10.12QRR93 pKa = 11.84NCYY96 pKa = 10.07RR97 pKa = 11.84EE98 pKa = 4.12VLRR101 pKa = 11.84GTAEE105 pKa = 4.04ALGEE109 pKa = 4.08AAHH112 pKa = 6.16TGAWGPLPMSPVPGPYY128 pKa = 10.18ARR130 pKa = 11.84GEE132 pKa = 4.08LDD134 pKa = 3.91SYY136 pKa = 11.1LPPPACRR143 pKa = 11.84RR144 pKa = 11.84LYY146 pKa = 10.28RR147 pKa = 11.84AQLYY151 pKa = 8.61HH152 pKa = 5.93QRR154 pKa = 11.84QVLLLYY160 pKa = 9.88LQYY163 pKa = 10.84YY164 pKa = 7.6PQVDD168 pKa = 3.99AGPPAGPP175 pKa = 3.83
II1 pKa = 7.28PKK3 pKa = 10.15GYY5 pKa = 8.95TIVRR9 pKa = 11.84GPLKK13 pKa = 10.66SIMRR17 pKa = 11.84TLQLKK22 pKa = 9.72QKK24 pKa = 9.13STLKK28 pKa = 10.69QMFGKK33 pKa = 10.1SCFKK37 pKa = 10.22TKK39 pKa = 10.5HH40 pKa = 5.74FLLSPAEE47 pKa = 4.08LPRR50 pKa = 11.84RR51 pKa = 11.84TPQSLLDD58 pKa = 3.93LAYY61 pKa = 10.36QAATSRR67 pKa = 11.84TAQQRR72 pKa = 11.84SPVYY76 pKa = 10.08FPLRR80 pKa = 11.84LPPCLLNSSLYY91 pKa = 10.12QRR93 pKa = 11.84NCYY96 pKa = 10.07RR97 pKa = 11.84EE98 pKa = 4.12VLRR101 pKa = 11.84GTAEE105 pKa = 4.04ALGEE109 pKa = 4.08AAHH112 pKa = 6.16TGAWGPLPMSPVPGPYY128 pKa = 10.18ARR130 pKa = 11.84GEE132 pKa = 4.08LDD134 pKa = 3.91SYY136 pKa = 11.1LPPPACRR143 pKa = 11.84RR144 pKa = 11.84LYY146 pKa = 10.28RR147 pKa = 11.84AQLYY151 pKa = 8.61HH152 pKa = 5.93QRR154 pKa = 11.84QVLLLYY160 pKa = 9.88LQYY163 pKa = 10.84YY164 pKa = 7.6PQVDD168 pKa = 3.99AGPPAGPP175 pKa = 3.83
Molecular weight: 19.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2456 |
93 |
621 |
350.9 |
39.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.067 ± 0.73 | 2.484 ± 0.562 |
5.7 ± 0.61 | 5.619 ± 0.4 |
3.379 ± 0.296 | 7.573 ± 1.106 |
2.239 ± 0.313 | 3.176 ± 0.262 |
4.56 ± 0.818 | 8.958 ± 0.709 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.954 ± 0.314 | 2.972 ± 0.378 |
7.085 ± 0.691 | 4.723 ± 0.558 |
6.678 ± 0.56 | 8.103 ± 0.55 |
5.863 ± 0.621 | 7.329 ± 0.705 |
1.792 ± 0.412 | 3.746 ± 0.39 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
