
Enhydrobacter aerosaccus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillales incertae sedis; Enhydrobacter
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
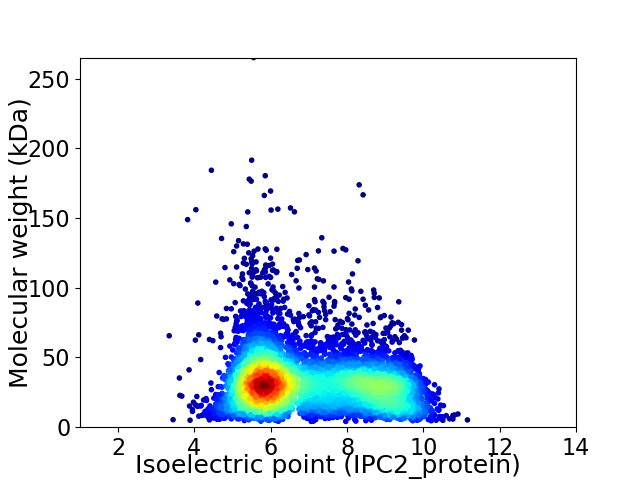
Virtual 2D-PAGE plot for 6507 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
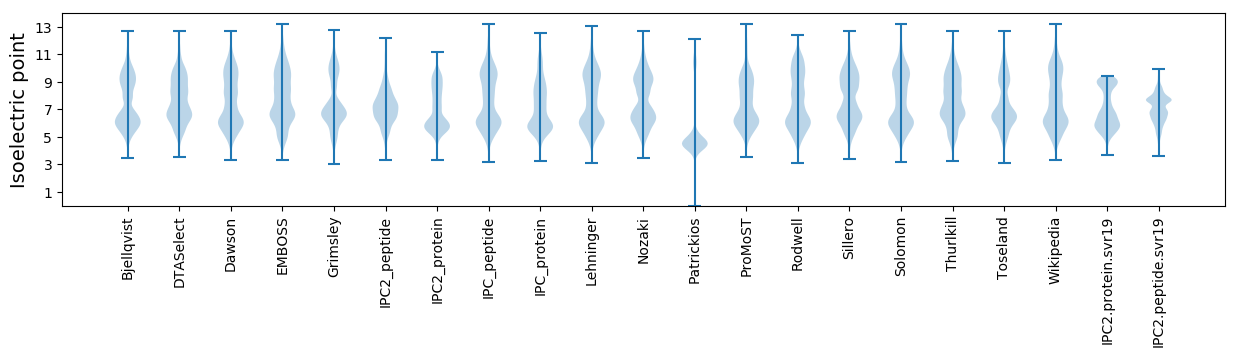
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1T4TH63|A0A1T4TH63_9PROT Serine--tRNA ligase OS=Enhydrobacter aerosaccus OX=225324 GN=serS PE=3 SV=1
MM1 pKa = 7.56IGGAGNDD8 pKa = 3.37TYY10 pKa = 11.75YY11 pKa = 11.1VDD13 pKa = 3.68NPGDD17 pKa = 3.59IVRR20 pKa = 11.84EE21 pKa = 3.94TAGAAFAVPAGWTLKK36 pKa = 9.3GTADD40 pKa = 3.6YY41 pKa = 11.4NGDD44 pKa = 3.59GEE46 pKa = 4.93LDD48 pKa = 3.75VVVSQGNTNQLWLLSGGALQSTVALPDD75 pKa = 3.77LSAGGWQFLGIADD88 pKa = 4.11EE89 pKa = 4.9NGDD92 pKa = 3.65GTKK95 pKa = 10.84DD96 pKa = 3.03LLYY99 pKa = 10.16RR100 pKa = 11.84HH101 pKa = 6.8TYY103 pKa = 9.27LGIYY107 pKa = 9.57YY108 pKa = 9.27SQLMNGATVGGGANAPVTTPDD129 pKa = 3.59PLQPLTASNQGNDD142 pKa = 3.55TVISSITYY150 pKa = 9.25ALTAGVEE157 pKa = 4.14NLTLAAGAGAIDD169 pKa = 3.51ATGNAVANLIVGNEE183 pKa = 3.88GNNRR187 pKa = 11.84ITGGGGADD195 pKa = 3.66TLTGGAGNDD204 pKa = 3.39TFVFTAASDD213 pKa = 3.97STPAAADD220 pKa = 3.83TITDD224 pKa = 4.5FTQGSDD230 pKa = 3.77SLDD233 pKa = 3.28LSAIGQFRR241 pKa = 11.84WLGTAAFDD249 pKa = 3.65HH250 pKa = 6.17QANALHH256 pKa = 6.1YY257 pKa = 9.59ASAGGITTLSADD269 pKa = 3.26INGDD273 pKa = 3.31GVADD277 pKa = 3.99FAVNLTGTLPLTTADD292 pKa = 3.71FTAASIAGASSAVMTPMDD310 pKa = 4.15AQSAQLIQAMATFDD324 pKa = 3.39TTAAGQSTLSAMPQDD339 pKa = 3.77STTIPLAVNHH349 pKa = 6.59HH350 pKa = 5.75NAA352 pKa = 3.55
MM1 pKa = 7.56IGGAGNDD8 pKa = 3.37TYY10 pKa = 11.75YY11 pKa = 11.1VDD13 pKa = 3.68NPGDD17 pKa = 3.59IVRR20 pKa = 11.84EE21 pKa = 3.94TAGAAFAVPAGWTLKK36 pKa = 9.3GTADD40 pKa = 3.6YY41 pKa = 11.4NGDD44 pKa = 3.59GEE46 pKa = 4.93LDD48 pKa = 3.75VVVSQGNTNQLWLLSGGALQSTVALPDD75 pKa = 3.77LSAGGWQFLGIADD88 pKa = 4.11EE89 pKa = 4.9NGDD92 pKa = 3.65GTKK95 pKa = 10.84DD96 pKa = 3.03LLYY99 pKa = 10.16RR100 pKa = 11.84HH101 pKa = 6.8TYY103 pKa = 9.27LGIYY107 pKa = 9.57YY108 pKa = 9.27SQLMNGATVGGGANAPVTTPDD129 pKa = 3.59PLQPLTASNQGNDD142 pKa = 3.55TVISSITYY150 pKa = 9.25ALTAGVEE157 pKa = 4.14NLTLAAGAGAIDD169 pKa = 3.51ATGNAVANLIVGNEE183 pKa = 3.88GNNRR187 pKa = 11.84ITGGGGADD195 pKa = 3.66TLTGGAGNDD204 pKa = 3.39TFVFTAASDD213 pKa = 3.97STPAAADD220 pKa = 3.83TITDD224 pKa = 4.5FTQGSDD230 pKa = 3.77SLDD233 pKa = 3.28LSAIGQFRR241 pKa = 11.84WLGTAAFDD249 pKa = 3.65HH250 pKa = 6.17QANALHH256 pKa = 6.1YY257 pKa = 9.59ASAGGITTLSADD269 pKa = 3.26INGDD273 pKa = 3.31GVADD277 pKa = 3.99FAVNLTGTLPLTTADD292 pKa = 3.71FTAASIAGASSAVMTPMDD310 pKa = 4.15AQSAQLIQAMATFDD324 pKa = 3.39TTAAGQSTLSAMPQDD339 pKa = 3.77STTIPLAVNHH349 pKa = 6.59HH350 pKa = 5.75NAA352 pKa = 3.55
Molecular weight: 35.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1T4QP33|A0A1T4QP33_9PROT Heme chaperone HemW OS=Enhydrobacter aerosaccus OX=225324 GN=SAMN02745126_03356 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.29QPSKK9 pKa = 10.24LIRR12 pKa = 11.84KK13 pKa = 8.44RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.45GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.33VIANRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84SGRR39 pKa = 11.84KK40 pKa = 8.96RR41 pKa = 11.84LSAA44 pKa = 4.09
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.29QPSKK9 pKa = 10.24LIRR12 pKa = 11.84KK13 pKa = 8.44RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.45GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.33VIANRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84SGRR39 pKa = 11.84KK40 pKa = 8.96RR41 pKa = 11.84LSAA44 pKa = 4.09
Molecular weight: 5.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2051812 |
39 |
2453 |
315.3 |
34.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.378 ± 0.037 | 0.888 ± 0.008 |
5.467 ± 0.024 | 5.267 ± 0.025 |
3.705 ± 0.02 | 8.645 ± 0.031 |
2.082 ± 0.015 | 4.951 ± 0.019 |
3.38 ± 0.025 | 10.317 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.46 ± 0.013 | 2.49 ± 0.017 |
5.508 ± 0.02 | 3.159 ± 0.016 |
7.313 ± 0.028 | 5.238 ± 0.022 |
5.283 ± 0.025 | 7.669 ± 0.03 |
1.481 ± 0.013 | 2.317 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
