
Enterobacteria phage P4 (Bacteriophage P4)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; unclassified Caudovirales
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
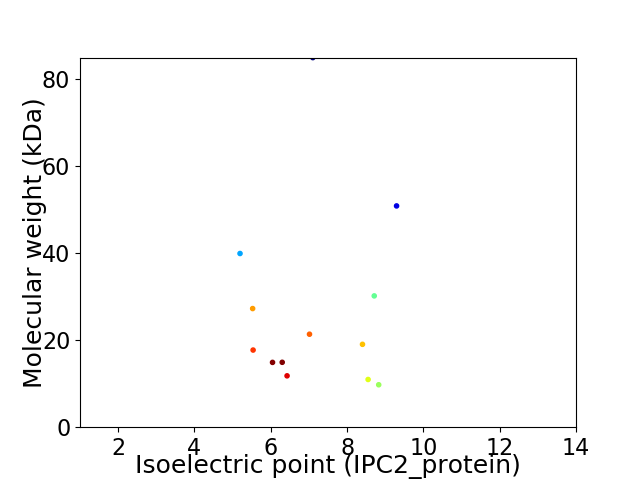
Virtual 2D-PAGE plot for 13 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P13058|GOP_BPP4 Protein gop OS=Enterobacteria phage P4 OX=10680 GN=gop PE=4 SV=1
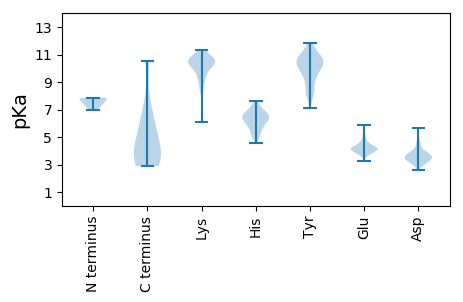
MM1 pKa = 7.31KK2 pKa = 10.34VYY4 pKa = 10.58FEE6 pKa = 4.53NYY8 pKa = 9.09SYY10 pKa = 11.56YY11 pKa = 9.67PALRR15 pKa = 11.84TRR17 pKa = 11.84SAEE20 pKa = 3.8MTGLNNLSYY29 pKa = 10.69EE30 pKa = 4.29NKK32 pKa = 10.29KK33 pKa = 10.67KK34 pKa = 10.27ILPLISLGKK43 pKa = 8.45WPRR46 pKa = 11.84SEE48 pKa = 4.55EE49 pKa = 3.98IQVSLDD55 pKa = 3.03KK56 pKa = 11.17SLEE59 pKa = 4.13VMSNLPFILDD69 pKa = 3.51VTKK72 pKa = 10.99DD73 pKa = 3.75NSHH76 pKa = 6.45HH77 pKa = 6.31CASSFEE83 pKa = 4.26LLSPEE88 pKa = 4.31NGFKK92 pKa = 10.6NWIEE96 pKa = 3.9FCSRR100 pKa = 11.84NDD102 pKa = 3.45NIIPVVQMPDD112 pKa = 3.08SAKK115 pKa = 10.63LRR117 pKa = 11.84DD118 pKa = 3.47ISIQARR124 pKa = 11.84VLEE127 pKa = 4.13EE128 pKa = 4.16LKK130 pKa = 11.0GSIAFRR136 pKa = 11.84IRR138 pKa = 11.84NLNTDD143 pKa = 2.94INKK146 pKa = 8.8TLTSLVSMNSPEE158 pKa = 3.94NAIVFIDD165 pKa = 3.68LGYY168 pKa = 10.41IRR170 pKa = 11.84GNVSAITAAAINSINQIRR188 pKa = 11.84TEE190 pKa = 3.82IPEE193 pKa = 4.49AIISVLATSFPSSVTNFCRR212 pKa = 11.84EE213 pKa = 3.99NGQSGYY219 pKa = 10.0IDD221 pKa = 3.54VIEE224 pKa = 5.2RR225 pKa = 11.84EE226 pKa = 4.08LHH228 pKa = 5.16QNIGGSDD235 pKa = 3.27VAIYY239 pKa = 10.36GDD241 pKa = 3.67HH242 pKa = 6.64GSIHH246 pKa = 6.19SVVYY250 pKa = 10.58DD251 pKa = 4.02NIIGRR256 pKa = 11.84YY257 pKa = 7.12VPRR260 pKa = 11.84IDD262 pKa = 4.55IALNDD267 pKa = 2.73SWYY270 pKa = 10.04FEE272 pKa = 4.64RR273 pKa = 11.84RR274 pKa = 11.84PGMNKK279 pKa = 9.95EE280 pKa = 4.11GFIEE284 pKa = 4.17AAKK287 pKa = 10.47SILAEE292 pKa = 3.94YY293 pKa = 9.46PHH295 pKa = 5.85YY296 pKa = 10.37QRR298 pKa = 11.84EE299 pKa = 4.36DD300 pKa = 2.88SWGAAMIRR308 pKa = 11.84NAAIGDD314 pKa = 3.78IAGMGSPAKK323 pKa = 9.75WIAVRR328 pKa = 11.84VNLHH332 pKa = 6.1LNKK335 pKa = 10.23QIEE338 pKa = 4.33LSEE341 pKa = 4.01ALQYY345 pKa = 11.48GFDD348 pKa = 4.07HH349 pKa = 7.62DD350 pKa = 4.69EE351 pKa = 4.06EE352 pKa = 5.18DD353 pKa = 5.07LII355 pKa = 6.09
MM1 pKa = 7.31KK2 pKa = 10.34VYY4 pKa = 10.58FEE6 pKa = 4.53NYY8 pKa = 9.09SYY10 pKa = 11.56YY11 pKa = 9.67PALRR15 pKa = 11.84TRR17 pKa = 11.84SAEE20 pKa = 3.8MTGLNNLSYY29 pKa = 10.69EE30 pKa = 4.29NKK32 pKa = 10.29KK33 pKa = 10.67KK34 pKa = 10.27ILPLISLGKK43 pKa = 8.45WPRR46 pKa = 11.84SEE48 pKa = 4.55EE49 pKa = 3.98IQVSLDD55 pKa = 3.03KK56 pKa = 11.17SLEE59 pKa = 4.13VMSNLPFILDD69 pKa = 3.51VTKK72 pKa = 10.99DD73 pKa = 3.75NSHH76 pKa = 6.45HH77 pKa = 6.31CASSFEE83 pKa = 4.26LLSPEE88 pKa = 4.31NGFKK92 pKa = 10.6NWIEE96 pKa = 3.9FCSRR100 pKa = 11.84NDD102 pKa = 3.45NIIPVVQMPDD112 pKa = 3.08SAKK115 pKa = 10.63LRR117 pKa = 11.84DD118 pKa = 3.47ISIQARR124 pKa = 11.84VLEE127 pKa = 4.13EE128 pKa = 4.16LKK130 pKa = 11.0GSIAFRR136 pKa = 11.84IRR138 pKa = 11.84NLNTDD143 pKa = 2.94INKK146 pKa = 8.8TLTSLVSMNSPEE158 pKa = 3.94NAIVFIDD165 pKa = 3.68LGYY168 pKa = 10.41IRR170 pKa = 11.84GNVSAITAAAINSINQIRR188 pKa = 11.84TEE190 pKa = 3.82IPEE193 pKa = 4.49AIISVLATSFPSSVTNFCRR212 pKa = 11.84EE213 pKa = 3.99NGQSGYY219 pKa = 10.0IDD221 pKa = 3.54VIEE224 pKa = 5.2RR225 pKa = 11.84EE226 pKa = 4.08LHH228 pKa = 5.16QNIGGSDD235 pKa = 3.27VAIYY239 pKa = 10.36GDD241 pKa = 3.67HH242 pKa = 6.64GSIHH246 pKa = 6.19SVVYY250 pKa = 10.58DD251 pKa = 4.02NIIGRR256 pKa = 11.84YY257 pKa = 7.12VPRR260 pKa = 11.84IDD262 pKa = 4.55IALNDD267 pKa = 2.73SWYY270 pKa = 10.04FEE272 pKa = 4.64RR273 pKa = 11.84RR274 pKa = 11.84PGMNKK279 pKa = 9.95EE280 pKa = 4.11GFIEE284 pKa = 4.17AAKK287 pKa = 10.47SILAEE292 pKa = 3.94YY293 pKa = 9.46PHH295 pKa = 5.85YY296 pKa = 10.37QRR298 pKa = 11.84EE299 pKa = 4.36DD300 pKa = 2.88SWGAAMIRR308 pKa = 11.84NAAIGDD314 pKa = 3.78IAGMGSPAKK323 pKa = 9.75WIAVRR328 pKa = 11.84VNLHH332 pKa = 6.1LNKK335 pKa = 10.23QIEE338 pKa = 4.33LSEE341 pKa = 4.01ALQYY345 pKa = 11.48GFDD348 pKa = 4.07HH349 pKa = 7.62DD350 pKa = 4.69EE351 pKa = 4.06EE352 pKa = 5.18DD353 pKa = 5.07LII355 pKa = 6.09
Molecular weight: 39.91 kDa
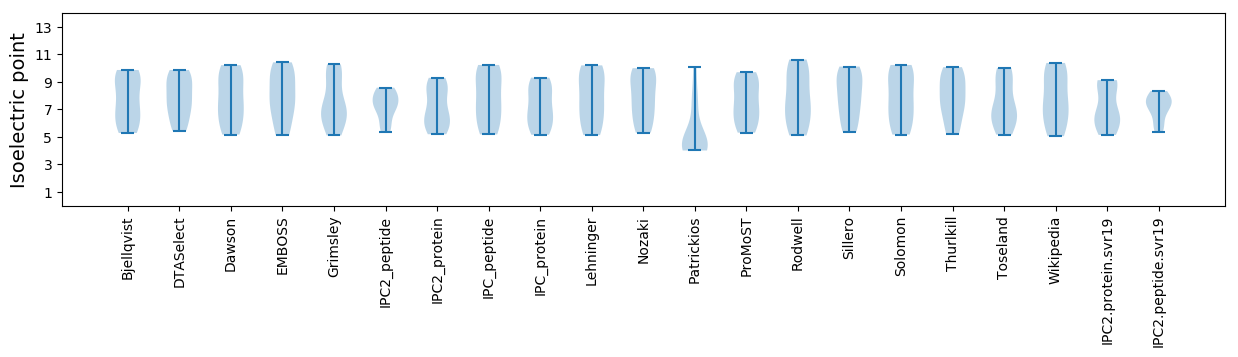
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P10277|PRIM_BPP4 Putative P4-specific DNA primase OS=Enterobacteria phage P4 OX=10680 GN=Alpha PE=1 SV=1
MM1 pKa = 7.52CPQMKK6 pKa = 10.39LNARR10 pKa = 11.84QVEE13 pKa = 4.31TAKK16 pKa = 10.91PKK18 pKa = 10.79DD19 pKa = 3.61KK20 pKa = 10.0TYY22 pKa = 11.59KK23 pKa = 9.33MADD26 pKa = 2.99GGGLYY31 pKa = 10.65LEE33 pKa = 5.0VSAKK37 pKa = 9.92GSKK40 pKa = 7.48YY41 pKa = 8.13WRR43 pKa = 11.84MKK45 pKa = 9.5YY46 pKa = 9.87RR47 pKa = 11.84RR48 pKa = 11.84PSDD51 pKa = 3.35KK52 pKa = 11.12KK53 pKa = 10.01EE54 pKa = 3.59DD55 pKa = 3.53RR56 pKa = 11.84LAFGVWPTVTLAQARR71 pKa = 11.84AKK73 pKa = 10.29RR74 pKa = 11.84DD75 pKa = 3.4EE76 pKa = 4.15AKK78 pKa = 10.53KK79 pKa = 10.84LLVQGIDD86 pKa = 3.19PKK88 pKa = 11.05VVQKK92 pKa = 9.3EE93 pKa = 3.86ARR95 pKa = 11.84AEE97 pKa = 3.8NSGAYY102 pKa = 8.54TFEE105 pKa = 5.91AIARR109 pKa = 11.84EE110 pKa = 3.81WHH112 pKa = 6.32ASNKK116 pKa = 9.2RR117 pKa = 11.84WSEE120 pKa = 3.87DD121 pKa = 2.76HH122 pKa = 6.69RR123 pKa = 11.84SRR125 pKa = 11.84VLRR128 pKa = 11.84YY129 pKa = 9.34LEE131 pKa = 5.45LYY133 pKa = 9.64IFPHH137 pKa = 6.58IGSSDD142 pKa = 2.76IRR144 pKa = 11.84QLKK147 pKa = 8.56TSHH150 pKa = 6.75LLAPIKK156 pKa = 10.64KK157 pKa = 8.48VDD159 pKa = 3.32ASGKK163 pKa = 9.46HH164 pKa = 6.14DD165 pKa = 3.36VAQRR169 pKa = 11.84LQQRR173 pKa = 11.84VTAIMRR179 pKa = 11.84YY180 pKa = 9.16AVQNDD185 pKa = 4.19YY186 pKa = 11.1IDD188 pKa = 4.69SNPASDD194 pKa = 3.41MAGALSTTKK203 pKa = 10.29ARR205 pKa = 11.84HH206 pKa = 5.04YY207 pKa = 9.05PALPSSRR214 pKa = 11.84FPEE217 pKa = 4.32FLARR221 pKa = 11.84LAAYY225 pKa = 9.53RR226 pKa = 11.84GRR228 pKa = 11.84VMTRR232 pKa = 11.84IAVKK236 pKa = 10.53LSLLTFVRR244 pKa = 11.84SSEE247 pKa = 3.82LRR249 pKa = 11.84FARR252 pKa = 11.84WDD254 pKa = 3.31EE255 pKa = 3.85FDD257 pKa = 3.71FDD259 pKa = 3.92KK260 pKa = 11.33SLWRR264 pKa = 11.84IPAKK268 pKa = 10.3RR269 pKa = 11.84EE270 pKa = 3.69EE271 pKa = 4.4IKK273 pKa = 10.66GVRR276 pKa = 11.84YY277 pKa = 9.94SYY279 pKa = 11.08RR280 pKa = 11.84GMKK283 pKa = 9.05MKK285 pKa = 10.39EE286 pKa = 3.7EE287 pKa = 4.71HH288 pKa = 6.46IVPLSRR294 pKa = 11.84QAMILLNQLKK304 pKa = 10.09QISGDD309 pKa = 3.64KK310 pKa = 10.47EE311 pKa = 3.96LLFPGDD317 pKa = 3.48HH318 pKa = 7.44DD319 pKa = 3.78ATKK322 pKa = 10.89VMSEE326 pKa = 3.95NTVNSALRR334 pKa = 11.84AMGYY338 pKa = 7.27DD339 pKa = 3.31TKK341 pKa = 11.12TEE343 pKa = 4.1VCGHH347 pKa = 6.36GFRR350 pKa = 11.84TMARR354 pKa = 11.84GALGEE359 pKa = 4.26SGLWSDD365 pKa = 5.4DD366 pKa = 3.84AIEE369 pKa = 4.4RR370 pKa = 11.84QLSHH374 pKa = 6.38SEE376 pKa = 4.1RR377 pKa = 11.84NNVRR381 pKa = 11.84AAYY384 pKa = 9.32IHH386 pKa = 5.95TSEE389 pKa = 4.67HH390 pKa = 5.72LDD392 pKa = 3.33EE393 pKa = 5.28RR394 pKa = 11.84RR395 pKa = 11.84LMMQWWADD403 pKa = 3.63YY404 pKa = 11.6LDD406 pKa = 3.77MNRR409 pKa = 11.84NKK411 pKa = 10.38YY412 pKa = 9.13ISLMIIQNTKK422 pKa = 10.48KK423 pKa = 10.35YY424 pKa = 10.5LNKK427 pKa = 10.15NSYY430 pKa = 8.32WLIFKK435 pKa = 10.07MSVKK439 pKa = 10.53
MM1 pKa = 7.52CPQMKK6 pKa = 10.39LNARR10 pKa = 11.84QVEE13 pKa = 4.31TAKK16 pKa = 10.91PKK18 pKa = 10.79DD19 pKa = 3.61KK20 pKa = 10.0TYY22 pKa = 11.59KK23 pKa = 9.33MADD26 pKa = 2.99GGGLYY31 pKa = 10.65LEE33 pKa = 5.0VSAKK37 pKa = 9.92GSKK40 pKa = 7.48YY41 pKa = 8.13WRR43 pKa = 11.84MKK45 pKa = 9.5YY46 pKa = 9.87RR47 pKa = 11.84RR48 pKa = 11.84PSDD51 pKa = 3.35KK52 pKa = 11.12KK53 pKa = 10.01EE54 pKa = 3.59DD55 pKa = 3.53RR56 pKa = 11.84LAFGVWPTVTLAQARR71 pKa = 11.84AKK73 pKa = 10.29RR74 pKa = 11.84DD75 pKa = 3.4EE76 pKa = 4.15AKK78 pKa = 10.53KK79 pKa = 10.84LLVQGIDD86 pKa = 3.19PKK88 pKa = 11.05VVQKK92 pKa = 9.3EE93 pKa = 3.86ARR95 pKa = 11.84AEE97 pKa = 3.8NSGAYY102 pKa = 8.54TFEE105 pKa = 5.91AIARR109 pKa = 11.84EE110 pKa = 3.81WHH112 pKa = 6.32ASNKK116 pKa = 9.2RR117 pKa = 11.84WSEE120 pKa = 3.87DD121 pKa = 2.76HH122 pKa = 6.69RR123 pKa = 11.84SRR125 pKa = 11.84VLRR128 pKa = 11.84YY129 pKa = 9.34LEE131 pKa = 5.45LYY133 pKa = 9.64IFPHH137 pKa = 6.58IGSSDD142 pKa = 2.76IRR144 pKa = 11.84QLKK147 pKa = 8.56TSHH150 pKa = 6.75LLAPIKK156 pKa = 10.64KK157 pKa = 8.48VDD159 pKa = 3.32ASGKK163 pKa = 9.46HH164 pKa = 6.14DD165 pKa = 3.36VAQRR169 pKa = 11.84LQQRR173 pKa = 11.84VTAIMRR179 pKa = 11.84YY180 pKa = 9.16AVQNDD185 pKa = 4.19YY186 pKa = 11.1IDD188 pKa = 4.69SNPASDD194 pKa = 3.41MAGALSTTKK203 pKa = 10.29ARR205 pKa = 11.84HH206 pKa = 5.04YY207 pKa = 9.05PALPSSRR214 pKa = 11.84FPEE217 pKa = 4.32FLARR221 pKa = 11.84LAAYY225 pKa = 9.53RR226 pKa = 11.84GRR228 pKa = 11.84VMTRR232 pKa = 11.84IAVKK236 pKa = 10.53LSLLTFVRR244 pKa = 11.84SSEE247 pKa = 3.82LRR249 pKa = 11.84FARR252 pKa = 11.84WDD254 pKa = 3.31EE255 pKa = 3.85FDD257 pKa = 3.71FDD259 pKa = 3.92KK260 pKa = 11.33SLWRR264 pKa = 11.84IPAKK268 pKa = 10.3RR269 pKa = 11.84EE270 pKa = 3.69EE271 pKa = 4.4IKK273 pKa = 10.66GVRR276 pKa = 11.84YY277 pKa = 9.94SYY279 pKa = 11.08RR280 pKa = 11.84GMKK283 pKa = 9.05MKK285 pKa = 10.39EE286 pKa = 3.7EE287 pKa = 4.71HH288 pKa = 6.46IVPLSRR294 pKa = 11.84QAMILLNQLKK304 pKa = 10.09QISGDD309 pKa = 3.64KK310 pKa = 10.47EE311 pKa = 3.96LLFPGDD317 pKa = 3.48HH318 pKa = 7.44DD319 pKa = 3.78ATKK322 pKa = 10.89VMSEE326 pKa = 3.95NTVNSALRR334 pKa = 11.84AMGYY338 pKa = 7.27DD339 pKa = 3.31TKK341 pKa = 11.12TEE343 pKa = 4.1VCGHH347 pKa = 6.36GFRR350 pKa = 11.84TMARR354 pKa = 11.84GALGEE359 pKa = 4.26SGLWSDD365 pKa = 5.4DD366 pKa = 3.84AIEE369 pKa = 4.4RR370 pKa = 11.84QLSHH374 pKa = 6.38SEE376 pKa = 4.1RR377 pKa = 11.84NNVRR381 pKa = 11.84AAYY384 pKa = 9.32IHH386 pKa = 5.95TSEE389 pKa = 4.67HH390 pKa = 5.72LDD392 pKa = 3.33EE393 pKa = 5.28RR394 pKa = 11.84RR395 pKa = 11.84LMMQWWADD403 pKa = 3.63YY404 pKa = 11.6LDD406 pKa = 3.77MNRR409 pKa = 11.84NKK411 pKa = 10.38YY412 pKa = 9.13ISLMIIQNTKK422 pKa = 10.48KK423 pKa = 10.35YY424 pKa = 10.5LNKK427 pKa = 10.15NSYY430 pKa = 8.32WLIFKK435 pKa = 10.07MSVKK439 pKa = 10.53
Molecular weight: 50.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3145 |
88 |
777 |
241.9 |
27.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.793 ± 0.945 | 1.494 ± 0.338 |
5.278 ± 0.472 | 6.455 ± 0.495 |
3.084 ± 0.354 | 5.723 ± 0.721 |
2.607 ± 0.316 | 5.564 ± 0.915 |
5.278 ± 0.662 | 9.539 ± 0.555 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.703 ± 0.301 | 4.102 ± 0.68 |
4.388 ± 0.696 | 3.879 ± 0.5 |
7.568 ± 0.562 | 7.154 ± 0.724 |
4.674 ± 0.605 | 6.041 ± 0.463 |
1.494 ± 0.194 | 3.18 ± 0.261 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
