
Human papillomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; Human papillomavirus types
Average proteome isoelectric point is 5.71
Get precalculated fractions of proteins
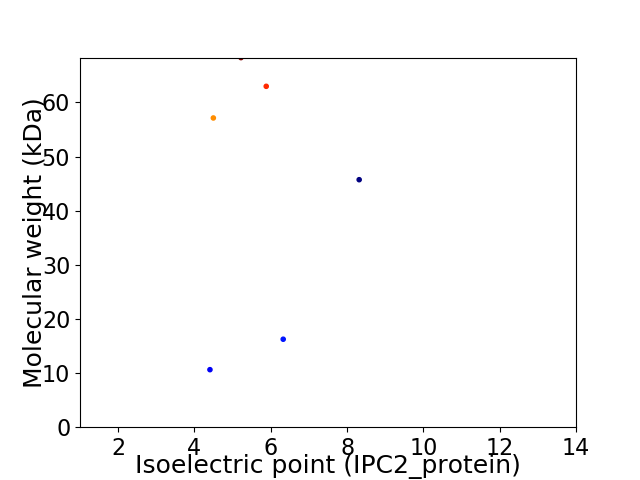
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
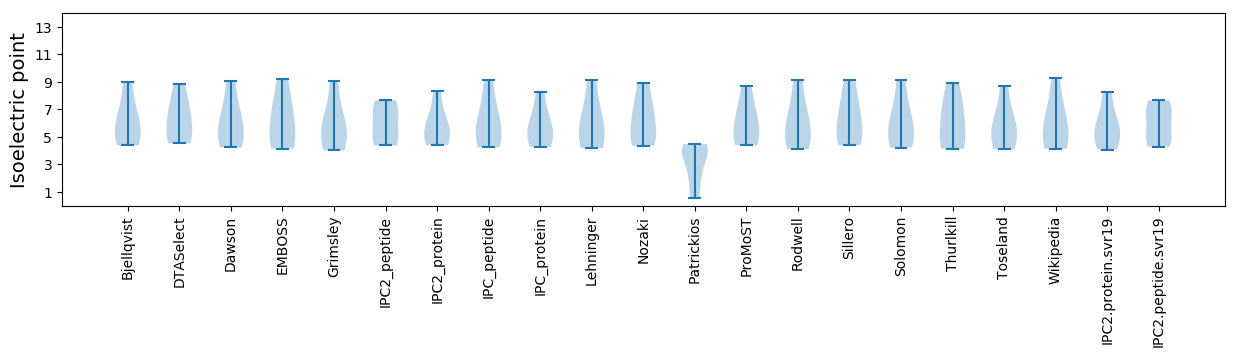
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1YWK4|A0A0K1YWK4_9PAPI Protein E6 OS=Human papillomavirus OX=10566 GN=E6 PE=3 SV=1
MM1 pKa = 7.67IGAQPDD7 pKa = 3.48IKK9 pKa = 11.1DD10 pKa = 3.28IDD12 pKa = 4.57LDD14 pKa = 3.88LQEE17 pKa = 5.75LVLPQNLLAEE27 pKa = 4.75EE28 pKa = 4.44SLSPDD33 pKa = 3.38ADD35 pKa = 3.72PEE37 pKa = 4.18EE38 pKa = 5.06EE39 pKa = 4.1EE40 pKa = 4.17QQLYY44 pKa = 8.01WVDD47 pKa = 3.93TCCGTCKK54 pKa = 10.06ATVRR58 pKa = 11.84VCVFATSTAVCTLQFLLQGQLSFVCILCSKK88 pKa = 9.62GRR90 pKa = 11.84RR91 pKa = 11.84HH92 pKa = 6.19GRR94 pKa = 11.84PNN96 pKa = 3.03
MM1 pKa = 7.67IGAQPDD7 pKa = 3.48IKK9 pKa = 11.1DD10 pKa = 3.28IDD12 pKa = 4.57LDD14 pKa = 3.88LQEE17 pKa = 5.75LVLPQNLLAEE27 pKa = 4.75EE28 pKa = 4.44SLSPDD33 pKa = 3.38ADD35 pKa = 3.72PEE37 pKa = 4.18EE38 pKa = 5.06EE39 pKa = 4.1EE40 pKa = 4.17QQLYY44 pKa = 8.01WVDD47 pKa = 3.93TCCGTCKK54 pKa = 10.06ATVRR58 pKa = 11.84VCVFATSTAVCTLQFLLQGQLSFVCILCSKK88 pKa = 9.62GRR90 pKa = 11.84RR91 pKa = 11.84HH92 pKa = 6.19GRR94 pKa = 11.84PNN96 pKa = 3.03
Molecular weight: 10.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1YX24|A0A0K1YX24_9PAPI Replication protein E1 OS=Human papillomavirus OX=10566 GN=E1 PE=3 SV=1
MM1 pKa = 7.04MRR3 pKa = 11.84MEE5 pKa = 4.21TQEE8 pKa = 4.05TLTDD12 pKa = 3.82RR13 pKa = 11.84FNALQDD19 pKa = 4.39AILNLIEE26 pKa = 4.98EE27 pKa = 4.66NPSDD31 pKa = 3.73LKK33 pKa = 11.31SIIRR37 pKa = 11.84YY38 pKa = 8.38WEE40 pKa = 3.99LNRR43 pKa = 11.84KK44 pKa = 7.44EE45 pKa = 4.97HH46 pKa = 5.0VTLYY50 pKa = 9.7YY51 pKa = 10.48ARR53 pKa = 11.84KK54 pKa = 9.74EE55 pKa = 3.98GFTRR59 pKa = 11.84LGLQPVPTPAVSEE72 pKa = 4.42YY73 pKa = 10.23NAKK76 pKa = 9.92QAIQMQLLVNSLSKK90 pKa = 10.88SVFAKK95 pKa = 7.6EE96 pKa = 3.57TWTLSDD102 pKa = 4.75CSAEE106 pKa = 5.05LINTLPKK113 pKa = 10.79DD114 pKa = 3.68CFKK117 pKa = 11.05KK118 pKa = 10.04QGYY121 pKa = 6.22TVEE124 pKa = 3.65VWYY127 pKa = 10.46DD128 pKa = 3.13NDD130 pKa = 3.65RR131 pKa = 11.84NKK133 pKa = 10.83AYY135 pKa = 9.57PYY137 pKa = 9.16TNWKK141 pKa = 9.36RR142 pKa = 11.84IYY144 pKa = 9.93YY145 pKa = 9.43QDD147 pKa = 4.75SHH149 pKa = 8.38DD150 pKa = 3.66EE151 pKa = 3.99WQLTEE156 pKa = 4.31GKK158 pKa = 10.12VDD160 pKa = 3.8EE161 pKa = 4.8NGLYY165 pKa = 10.31YY166 pKa = 10.31DD167 pKa = 4.39EE168 pKa = 5.56KK169 pKa = 11.35NGDD172 pKa = 3.08RR173 pKa = 11.84VYY175 pKa = 11.47FMLFGADD182 pKa = 3.1ADD184 pKa = 4.11KK185 pKa = 11.67YY186 pKa = 11.0GVTKK190 pKa = 10.09EE191 pKa = 4.03WTVHH195 pKa = 5.38YY196 pKa = 10.02RR197 pKa = 11.84DD198 pKa = 3.59TTIVSSSSSSRR209 pKa = 11.84RR210 pKa = 11.84VADD213 pKa = 3.2SSQAFTQPSTSGYY226 pKa = 10.69SKK228 pKa = 9.69TPSPKK233 pKa = 9.02RR234 pKa = 11.84RR235 pKa = 11.84RR236 pKa = 11.84AAEE239 pKa = 3.96EE240 pKa = 4.21ADD242 pKa = 3.59SSTSSSSSSAPSGSPASNLRR262 pKa = 11.84DD263 pKa = 3.28RR264 pKa = 11.84GGGQGEE270 pKa = 4.06RR271 pKa = 11.84SVPLSKK277 pKa = 10.27RR278 pKa = 11.84RR279 pKa = 11.84RR280 pKa = 11.84HH281 pKa = 4.92TSEE284 pKa = 3.82SAGEE288 pKa = 4.2SAVPSPSEE296 pKa = 3.75VGTGHH301 pKa = 7.75RR302 pKa = 11.84LLARR306 pKa = 11.84QALGRR311 pKa = 11.84VQRR314 pKa = 11.84LQAEE318 pKa = 4.1AWDD321 pKa = 4.03PFILILKK328 pKa = 9.29GPSNNLKK335 pKa = 10.14CWRR338 pKa = 11.84NRR340 pKa = 11.84NKK342 pKa = 10.29HH343 pKa = 4.34NHH345 pKa = 4.08VHH347 pKa = 5.89VKK349 pKa = 10.45ASSNVWRR356 pKa = 11.84WLGEE360 pKa = 3.88GYY362 pKa = 9.2TYY364 pKa = 11.52SRR366 pKa = 11.84MLLAFDD372 pKa = 4.55SEE374 pKa = 4.81TEE376 pKa = 3.9RR377 pKa = 11.84EE378 pKa = 4.0RR379 pKa = 11.84FVQFTRR385 pKa = 11.84FPKK388 pKa = 9.88DD389 pKa = 2.81TTYY392 pKa = 11.19AYY394 pKa = 10.76GNLEE398 pKa = 4.04SLL400 pKa = 4.22
MM1 pKa = 7.04MRR3 pKa = 11.84MEE5 pKa = 4.21TQEE8 pKa = 4.05TLTDD12 pKa = 3.82RR13 pKa = 11.84FNALQDD19 pKa = 4.39AILNLIEE26 pKa = 4.98EE27 pKa = 4.66NPSDD31 pKa = 3.73LKK33 pKa = 11.31SIIRR37 pKa = 11.84YY38 pKa = 8.38WEE40 pKa = 3.99LNRR43 pKa = 11.84KK44 pKa = 7.44EE45 pKa = 4.97HH46 pKa = 5.0VTLYY50 pKa = 9.7YY51 pKa = 10.48ARR53 pKa = 11.84KK54 pKa = 9.74EE55 pKa = 3.98GFTRR59 pKa = 11.84LGLQPVPTPAVSEE72 pKa = 4.42YY73 pKa = 10.23NAKK76 pKa = 9.92QAIQMQLLVNSLSKK90 pKa = 10.88SVFAKK95 pKa = 7.6EE96 pKa = 3.57TWTLSDD102 pKa = 4.75CSAEE106 pKa = 5.05LINTLPKK113 pKa = 10.79DD114 pKa = 3.68CFKK117 pKa = 11.05KK118 pKa = 10.04QGYY121 pKa = 6.22TVEE124 pKa = 3.65VWYY127 pKa = 10.46DD128 pKa = 3.13NDD130 pKa = 3.65RR131 pKa = 11.84NKK133 pKa = 10.83AYY135 pKa = 9.57PYY137 pKa = 9.16TNWKK141 pKa = 9.36RR142 pKa = 11.84IYY144 pKa = 9.93YY145 pKa = 9.43QDD147 pKa = 4.75SHH149 pKa = 8.38DD150 pKa = 3.66EE151 pKa = 3.99WQLTEE156 pKa = 4.31GKK158 pKa = 10.12VDD160 pKa = 3.8EE161 pKa = 4.8NGLYY165 pKa = 10.31YY166 pKa = 10.31DD167 pKa = 4.39EE168 pKa = 5.56KK169 pKa = 11.35NGDD172 pKa = 3.08RR173 pKa = 11.84VYY175 pKa = 11.47FMLFGADD182 pKa = 3.1ADD184 pKa = 4.11KK185 pKa = 11.67YY186 pKa = 11.0GVTKK190 pKa = 10.09EE191 pKa = 4.03WTVHH195 pKa = 5.38YY196 pKa = 10.02RR197 pKa = 11.84DD198 pKa = 3.59TTIVSSSSSSRR209 pKa = 11.84RR210 pKa = 11.84VADD213 pKa = 3.2SSQAFTQPSTSGYY226 pKa = 10.69SKK228 pKa = 9.69TPSPKK233 pKa = 9.02RR234 pKa = 11.84RR235 pKa = 11.84RR236 pKa = 11.84AAEE239 pKa = 3.96EE240 pKa = 4.21ADD242 pKa = 3.59SSTSSSSSSAPSGSPASNLRR262 pKa = 11.84DD263 pKa = 3.28RR264 pKa = 11.84GGGQGEE270 pKa = 4.06RR271 pKa = 11.84SVPLSKK277 pKa = 10.27RR278 pKa = 11.84RR279 pKa = 11.84RR280 pKa = 11.84HH281 pKa = 4.92TSEE284 pKa = 3.82SAGEE288 pKa = 4.2SAVPSPSEE296 pKa = 3.75VGTGHH301 pKa = 7.75RR302 pKa = 11.84LLARR306 pKa = 11.84QALGRR311 pKa = 11.84VQRR314 pKa = 11.84LQAEE318 pKa = 4.1AWDD321 pKa = 4.03PFILILKK328 pKa = 9.29GPSNNLKK335 pKa = 10.14CWRR338 pKa = 11.84NRR340 pKa = 11.84NKK342 pKa = 10.29HH343 pKa = 4.34NHH345 pKa = 4.08VHH347 pKa = 5.89VKK349 pKa = 10.45ASSNVWRR356 pKa = 11.84WLGEE360 pKa = 3.88GYY362 pKa = 9.2TYY364 pKa = 11.52SRR366 pKa = 11.84MLLAFDD372 pKa = 4.55SEE374 pKa = 4.81TEE376 pKa = 3.9RR377 pKa = 11.84EE378 pKa = 4.0RR379 pKa = 11.84FVQFTRR385 pKa = 11.84FPKK388 pKa = 9.88DD389 pKa = 2.81TTYY392 pKa = 11.19AYY394 pKa = 10.76GNLEE398 pKa = 4.04SLL400 pKa = 4.22
Molecular weight: 45.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2309 |
96 |
600 |
384.8 |
43.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.933 ± 0.319 | 2.252 ± 0.92 |
6.063 ± 0.315 | 6.54 ± 0.563 |
4.807 ± 0.471 | 5.024 ± 0.562 |
1.689 ± 0.169 | 4.894 ± 1.072 |
4.981 ± 0.811 | 9.181 ± 0.61 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.602 ± 0.382 | 5.24 ± 0.372 |
5.673 ± 1.148 | 4.721 ± 0.365 |
5.37 ± 0.69 | 8.922 ± 0.651 |
5.544 ± 0.699 | 6.453 ± 0.33 |
1.343 ± 0.404 | 3.768 ± 0.408 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
