
Nostoc sp. 3335mG
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Nostocales; Nostocaceae; Nostoc; unclassified Nostoc
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
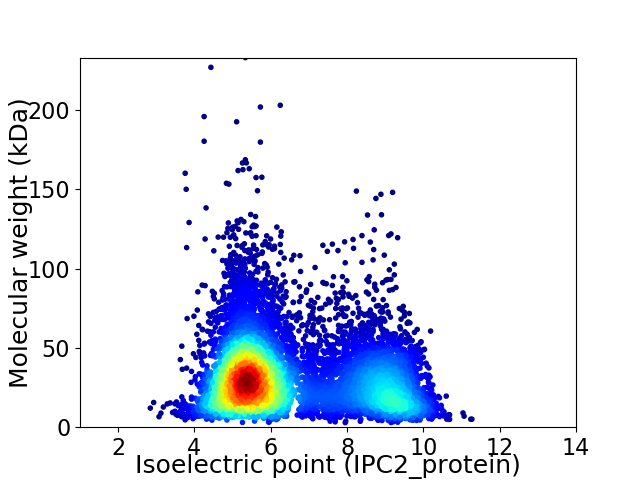
Virtual 2D-PAGE plot for 8647 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A318BD35|A0A318BD35_9NOSO Uncharacterized protein OS=Nostoc sp. 3335mG OX=2204170 GN=DMC47_14130 PE=4 SV=1
MM1 pKa = 7.39TKK3 pKa = 10.38LAKK6 pKa = 10.01TGDD9 pKa = 3.7GVSRR13 pKa = 11.84RR14 pKa = 11.84LLSGTALASAAACALIATPVYY35 pKa = 10.57AQDD38 pKa = 3.33EE39 pKa = 4.7CGPTTEE45 pKa = 4.46GTVTCTAANNPYY57 pKa = 9.97PNGIEE62 pKa = 4.11YY63 pKa = 7.6STPAVDD69 pKa = 4.02PLQDD73 pKa = 4.23PGLDD77 pKa = 3.67PNAPVYY83 pKa = 10.89DD84 pKa = 3.66LTVNLGSGVAIDD96 pKa = 4.9ADD98 pKa = 3.64ATRR101 pKa = 11.84PGVALIGFNDD111 pKa = 4.21GAVTLNSFGDD121 pKa = 3.54TSITVTGVGAQGVLASTYY139 pKa = 10.22TGDD142 pKa = 3.61LTISTDD148 pKa = 3.47SITATGGASGGINANSSAGTITINADD174 pKa = 3.43TTSVSGNGSAGISANSFAGNVDD196 pKa = 2.86ITAGTVTAEE205 pKa = 4.26GLASSGIVAYY215 pKa = 10.67SNQGDD220 pKa = 3.77VTINAGTVTTTADD233 pKa = 2.91GGYY236 pKa = 10.45FGINSAAIEE245 pKa = 4.08AVSVQGTTNVTVDD258 pKa = 3.49SVSTTGVYY266 pKa = 10.19AAGISAYY273 pKa = 10.36SGVSANVTADD283 pKa = 3.83TISTTGYY290 pKa = 9.63ASYY293 pKa = 10.84GAIVQSRR300 pKa = 11.84GDD302 pKa = 3.22ASLTVGSISTAGDD315 pKa = 3.56YY316 pKa = 9.42STGAFVQTLNGTASLTSTGSITTTGDD342 pKa = 3.08DD343 pKa = 4.12AIGVFVDD350 pKa = 4.59SVAGNATVVVNDD362 pKa = 3.56VSTTGDD368 pKa = 3.02NSTAVYY374 pKa = 10.88ADD376 pKa = 3.49GDD378 pKa = 4.06IVDD381 pKa = 3.71VTINGDD387 pKa = 3.15VSTQGLASFGVDD399 pKa = 2.89AFGTDD404 pKa = 3.17TVTVTNNGTITTAGDD419 pKa = 3.46GSIGVLVTGVNGAVVDD435 pKa = 4.45GSGSVSTSGDD445 pKa = 3.01NSRR448 pKa = 11.84GLSIRR453 pKa = 11.84SVFGDD458 pKa = 3.23ADD460 pKa = 3.8VTVGDD465 pKa = 4.03VTTTGANSAAVVATSDD481 pKa = 3.7FGNVSVTNTGAIDD494 pKa = 3.65TSGDD498 pKa = 2.99ASAGIRR504 pKa = 11.84AVAFTGNATVANNGTVTTTGLGSTGIYY531 pKa = 9.86AGAYY535 pKa = 9.79GDD537 pKa = 3.82VTVTGAGSVASSGVGIEE554 pKa = 4.41AYY556 pKa = 10.27SVAGSIDD563 pKa = 3.34ITHH566 pKa = 6.95GDD568 pKa = 3.07IATTEE573 pKa = 4.41DD574 pKa = 3.74GASGIVAEE582 pKa = 4.48TGNYY586 pKa = 9.8NYY588 pKa = 10.82AAGNITIDD596 pKa = 3.56VGDD599 pKa = 3.65ITTTGDD605 pKa = 3.25NADD608 pKa = 4.88GINASALSGGTIDD621 pKa = 3.86ITHH624 pKa = 6.78GAITTSGTNAFGVFAIGVDD643 pKa = 3.76DD644 pKa = 4.21VSISGTSATTTGEE657 pKa = 4.06DD658 pKa = 3.11AAGVYY663 pKa = 9.81GVSLFGDD670 pKa = 3.48VTIDD674 pKa = 3.14VGTVSTAGEE683 pKa = 4.13YY684 pKa = 10.98APGVVGAALLGGDD697 pKa = 3.73VSITAGTITTTGDD710 pKa = 3.38TSKK713 pKa = 11.0GAYY716 pKa = 9.2GLSRR720 pKa = 11.84YY721 pKa = 9.63GSTAITVGNVTTSGDD736 pKa = 3.45YY737 pKa = 10.99SIGVDD742 pKa = 3.37AYY744 pKa = 9.63AYY746 pKa = 9.34YY747 pKa = 10.3GASVTVNGTVATSGDD762 pKa = 3.84YY763 pKa = 11.04APGVNVYY770 pKa = 10.55AVGLGGTDD778 pKa = 2.8VDD780 pKa = 4.3NNATVVNNGAIRR792 pKa = 11.84TSGIEE797 pKa = 4.1SNGITAEE804 pKa = 4.04AVFGDD809 pKa = 4.01VVISGTGSVATTGDD823 pKa = 3.58FSTGILARR831 pKa = 11.84SANGVVDD838 pKa = 3.73ITAGTVTTTGIASGIDD854 pKa = 3.41AYY856 pKa = 10.91ASGGVSITVDD866 pKa = 3.42TVRR869 pKa = 11.84TAGADD874 pKa = 3.5SNAINAGGFYY884 pKa = 10.64SVDD887 pKa = 3.09VTANTVSTTGASANGIDD904 pKa = 3.45AVSFGGDD911 pKa = 3.22VTVAAGSVIASGADD925 pKa = 3.22STAIRR930 pKa = 11.84ALGFGGGANVSVTGAVRR947 pKa = 11.84STSGTAIDD955 pKa = 4.21ILAAGAGGAGGPAVGDD971 pKa = 3.69PANDD975 pKa = 4.74GIARR979 pKa = 11.84VLVGTNGSVQGGTNAITTDD998 pKa = 3.88ALNGTQITNRR1008 pKa = 11.84GSIIGGSGYY1017 pKa = 10.64AIEE1020 pKa = 4.71ASGGAATINNIGSITGRR1037 pKa = 11.84LLLTDD1042 pKa = 4.01NSDD1045 pKa = 3.43TLTNSGNFTLIGNSDD1060 pKa = 3.57FGGGNDD1066 pKa = 4.27LLTNSGTVRR1075 pKa = 11.84FGSAAGTQNITLVGLEE1091 pKa = 4.11SFANSGLVDD1100 pKa = 4.11LRR1102 pKa = 11.84NGVAGDD1108 pKa = 3.86RR1109 pKa = 11.84LTLAAYY1115 pKa = 7.59TGSGPAALGLDD1126 pKa = 3.39IAFGATPTVDD1136 pKa = 2.96RR1137 pKa = 11.84LNVTTATGSTSVVLNPLNGPATLIPTTTLIQASAASSATAFTLSAGSQFNGLIQYY1192 pKa = 9.62GLVYY1196 pKa = 10.67NPTSFAYY1203 pKa = 10.24QLISAPNATVYY1214 pKa = 10.42RR1215 pKa = 11.84QAKK1218 pKa = 8.73LGEE1221 pKa = 4.78GISTLWNRR1229 pKa = 11.84SADD1232 pKa = 3.81AITAHH1237 pKa = 6.94LAADD1241 pKa = 4.44RR1242 pKa = 11.84DD1243 pKa = 4.6SGWSAPTADD1252 pKa = 3.24SAGRR1256 pKa = 11.84IWLQMFGEE1264 pKa = 4.42VNKK1267 pKa = 10.32RR1268 pKa = 11.84DD1269 pKa = 3.57EE1270 pKa = 4.23TRR1272 pKa = 11.84SFGFNGLAQNDD1283 pKa = 3.06VDD1285 pKa = 5.45LGYY1288 pKa = 10.57KK1289 pKa = 9.77QDD1291 pKa = 3.46VFGGQIGFDD1300 pKa = 3.61LYY1302 pKa = 10.41GTRR1305 pKa = 11.84SDD1307 pKa = 3.28TGGFTVGVSGGYY1319 pKa = 9.95ASSVMHH1325 pKa = 6.41FAGNDD1330 pKa = 3.06RR1331 pKa = 11.84FQIDD1335 pKa = 3.81AVNGTLYY1342 pKa = 10.95AGFRR1346 pKa = 11.84AGGLFLNGLAKK1357 pKa = 10.23YY1358 pKa = 10.29DD1359 pKa = 3.78YY1360 pKa = 11.46AWIKK1364 pKa = 10.53NRR1366 pKa = 11.84GNAPVSTFNLEE1377 pKa = 4.03TKK1379 pKa = 9.68AQTWGGKK1386 pKa = 9.26VEE1388 pKa = 4.01AGFRR1392 pKa = 11.84FGSNSFFIEE1401 pKa = 3.84PAASIAYY1408 pKa = 7.56TSTDD1412 pKa = 2.69MDD1414 pKa = 3.35QYY1416 pKa = 11.42GVYY1419 pKa = 10.59GGTFDD1424 pKa = 5.8FDD1426 pKa = 4.47DD1427 pKa = 3.99FTGLRR1432 pKa = 11.84GKK1434 pKa = 10.44AGARR1438 pKa = 11.84IGGSTPVGSSTLVFYY1453 pKa = 11.09AGAAAVHH1460 pKa = 5.79EE1461 pKa = 4.88FKK1463 pKa = 11.44GEE1465 pKa = 4.08DD1466 pKa = 3.39GLLFSSGGQTVRR1478 pKa = 11.84LTNDD1482 pKa = 2.82RR1483 pKa = 11.84LGTYY1487 pKa = 9.8GQGTLGLNIVTSSGVTGFIEE1507 pKa = 4.36AHH1509 pKa = 5.91GEE1511 pKa = 4.13YY1512 pKa = 10.13NDD1514 pKa = 4.11EE1515 pKa = 4.15YY1516 pKa = 11.34SGGGGRR1522 pKa = 11.84AGIRR1526 pKa = 11.84IKK1528 pKa = 10.8FF1529 pKa = 3.53
MM1 pKa = 7.39TKK3 pKa = 10.38LAKK6 pKa = 10.01TGDD9 pKa = 3.7GVSRR13 pKa = 11.84RR14 pKa = 11.84LLSGTALASAAACALIATPVYY35 pKa = 10.57AQDD38 pKa = 3.33EE39 pKa = 4.7CGPTTEE45 pKa = 4.46GTVTCTAANNPYY57 pKa = 9.97PNGIEE62 pKa = 4.11YY63 pKa = 7.6STPAVDD69 pKa = 4.02PLQDD73 pKa = 4.23PGLDD77 pKa = 3.67PNAPVYY83 pKa = 10.89DD84 pKa = 3.66LTVNLGSGVAIDD96 pKa = 4.9ADD98 pKa = 3.64ATRR101 pKa = 11.84PGVALIGFNDD111 pKa = 4.21GAVTLNSFGDD121 pKa = 3.54TSITVTGVGAQGVLASTYY139 pKa = 10.22TGDD142 pKa = 3.61LTISTDD148 pKa = 3.47SITATGGASGGINANSSAGTITINADD174 pKa = 3.43TTSVSGNGSAGISANSFAGNVDD196 pKa = 2.86ITAGTVTAEE205 pKa = 4.26GLASSGIVAYY215 pKa = 10.67SNQGDD220 pKa = 3.77VTINAGTVTTTADD233 pKa = 2.91GGYY236 pKa = 10.45FGINSAAIEE245 pKa = 4.08AVSVQGTTNVTVDD258 pKa = 3.49SVSTTGVYY266 pKa = 10.19AAGISAYY273 pKa = 10.36SGVSANVTADD283 pKa = 3.83TISTTGYY290 pKa = 9.63ASYY293 pKa = 10.84GAIVQSRR300 pKa = 11.84GDD302 pKa = 3.22ASLTVGSISTAGDD315 pKa = 3.56YY316 pKa = 9.42STGAFVQTLNGTASLTSTGSITTTGDD342 pKa = 3.08DD343 pKa = 4.12AIGVFVDD350 pKa = 4.59SVAGNATVVVNDD362 pKa = 3.56VSTTGDD368 pKa = 3.02NSTAVYY374 pKa = 10.88ADD376 pKa = 3.49GDD378 pKa = 4.06IVDD381 pKa = 3.71VTINGDD387 pKa = 3.15VSTQGLASFGVDD399 pKa = 2.89AFGTDD404 pKa = 3.17TVTVTNNGTITTAGDD419 pKa = 3.46GSIGVLVTGVNGAVVDD435 pKa = 4.45GSGSVSTSGDD445 pKa = 3.01NSRR448 pKa = 11.84GLSIRR453 pKa = 11.84SVFGDD458 pKa = 3.23ADD460 pKa = 3.8VTVGDD465 pKa = 4.03VTTTGANSAAVVATSDD481 pKa = 3.7FGNVSVTNTGAIDD494 pKa = 3.65TSGDD498 pKa = 2.99ASAGIRR504 pKa = 11.84AVAFTGNATVANNGTVTTTGLGSTGIYY531 pKa = 9.86AGAYY535 pKa = 9.79GDD537 pKa = 3.82VTVTGAGSVASSGVGIEE554 pKa = 4.41AYY556 pKa = 10.27SVAGSIDD563 pKa = 3.34ITHH566 pKa = 6.95GDD568 pKa = 3.07IATTEE573 pKa = 4.41DD574 pKa = 3.74GASGIVAEE582 pKa = 4.48TGNYY586 pKa = 9.8NYY588 pKa = 10.82AAGNITIDD596 pKa = 3.56VGDD599 pKa = 3.65ITTTGDD605 pKa = 3.25NADD608 pKa = 4.88GINASALSGGTIDD621 pKa = 3.86ITHH624 pKa = 6.78GAITTSGTNAFGVFAIGVDD643 pKa = 3.76DD644 pKa = 4.21VSISGTSATTTGEE657 pKa = 4.06DD658 pKa = 3.11AAGVYY663 pKa = 9.81GVSLFGDD670 pKa = 3.48VTIDD674 pKa = 3.14VGTVSTAGEE683 pKa = 4.13YY684 pKa = 10.98APGVVGAALLGGDD697 pKa = 3.73VSITAGTITTTGDD710 pKa = 3.38TSKK713 pKa = 11.0GAYY716 pKa = 9.2GLSRR720 pKa = 11.84YY721 pKa = 9.63GSTAITVGNVTTSGDD736 pKa = 3.45YY737 pKa = 10.99SIGVDD742 pKa = 3.37AYY744 pKa = 9.63AYY746 pKa = 9.34YY747 pKa = 10.3GASVTVNGTVATSGDD762 pKa = 3.84YY763 pKa = 11.04APGVNVYY770 pKa = 10.55AVGLGGTDD778 pKa = 2.8VDD780 pKa = 4.3NNATVVNNGAIRR792 pKa = 11.84TSGIEE797 pKa = 4.1SNGITAEE804 pKa = 4.04AVFGDD809 pKa = 4.01VVISGTGSVATTGDD823 pKa = 3.58FSTGILARR831 pKa = 11.84SANGVVDD838 pKa = 3.73ITAGTVTTTGIASGIDD854 pKa = 3.41AYY856 pKa = 10.91ASGGVSITVDD866 pKa = 3.42TVRR869 pKa = 11.84TAGADD874 pKa = 3.5SNAINAGGFYY884 pKa = 10.64SVDD887 pKa = 3.09VTANTVSTTGASANGIDD904 pKa = 3.45AVSFGGDD911 pKa = 3.22VTVAAGSVIASGADD925 pKa = 3.22STAIRR930 pKa = 11.84ALGFGGGANVSVTGAVRR947 pKa = 11.84STSGTAIDD955 pKa = 4.21ILAAGAGGAGGPAVGDD971 pKa = 3.69PANDD975 pKa = 4.74GIARR979 pKa = 11.84VLVGTNGSVQGGTNAITTDD998 pKa = 3.88ALNGTQITNRR1008 pKa = 11.84GSIIGGSGYY1017 pKa = 10.64AIEE1020 pKa = 4.71ASGGAATINNIGSITGRR1037 pKa = 11.84LLLTDD1042 pKa = 4.01NSDD1045 pKa = 3.43TLTNSGNFTLIGNSDD1060 pKa = 3.57FGGGNDD1066 pKa = 4.27LLTNSGTVRR1075 pKa = 11.84FGSAAGTQNITLVGLEE1091 pKa = 4.11SFANSGLVDD1100 pKa = 4.11LRR1102 pKa = 11.84NGVAGDD1108 pKa = 3.86RR1109 pKa = 11.84LTLAAYY1115 pKa = 7.59TGSGPAALGLDD1126 pKa = 3.39IAFGATPTVDD1136 pKa = 2.96RR1137 pKa = 11.84LNVTTATGSTSVVLNPLNGPATLIPTTTLIQASAASSATAFTLSAGSQFNGLIQYY1192 pKa = 9.62GLVYY1196 pKa = 10.67NPTSFAYY1203 pKa = 10.24QLISAPNATVYY1214 pKa = 10.42RR1215 pKa = 11.84QAKK1218 pKa = 8.73LGEE1221 pKa = 4.78GISTLWNRR1229 pKa = 11.84SADD1232 pKa = 3.81AITAHH1237 pKa = 6.94LAADD1241 pKa = 4.44RR1242 pKa = 11.84DD1243 pKa = 4.6SGWSAPTADD1252 pKa = 3.24SAGRR1256 pKa = 11.84IWLQMFGEE1264 pKa = 4.42VNKK1267 pKa = 10.32RR1268 pKa = 11.84DD1269 pKa = 3.57EE1270 pKa = 4.23TRR1272 pKa = 11.84SFGFNGLAQNDD1283 pKa = 3.06VDD1285 pKa = 5.45LGYY1288 pKa = 10.57KK1289 pKa = 9.77QDD1291 pKa = 3.46VFGGQIGFDD1300 pKa = 3.61LYY1302 pKa = 10.41GTRR1305 pKa = 11.84SDD1307 pKa = 3.28TGGFTVGVSGGYY1319 pKa = 9.95ASSVMHH1325 pKa = 6.41FAGNDD1330 pKa = 3.06RR1331 pKa = 11.84FQIDD1335 pKa = 3.81AVNGTLYY1342 pKa = 10.95AGFRR1346 pKa = 11.84AGGLFLNGLAKK1357 pKa = 10.23YY1358 pKa = 10.29DD1359 pKa = 3.78YY1360 pKa = 11.46AWIKK1364 pKa = 10.53NRR1366 pKa = 11.84GNAPVSTFNLEE1377 pKa = 4.03TKK1379 pKa = 9.68AQTWGGKK1386 pKa = 9.26VEE1388 pKa = 4.01AGFRR1392 pKa = 11.84FGSNSFFIEE1401 pKa = 3.84PAASIAYY1408 pKa = 7.56TSTDD1412 pKa = 2.69MDD1414 pKa = 3.35QYY1416 pKa = 11.42GVYY1419 pKa = 10.59GGTFDD1424 pKa = 5.8FDD1426 pKa = 4.47DD1427 pKa = 3.99FTGLRR1432 pKa = 11.84GKK1434 pKa = 10.44AGARR1438 pKa = 11.84IGGSTPVGSSTLVFYY1453 pKa = 11.09AGAAAVHH1460 pKa = 5.79EE1461 pKa = 4.88FKK1463 pKa = 11.44GEE1465 pKa = 4.08DD1466 pKa = 3.39GLLFSSGGQTVRR1478 pKa = 11.84LTNDD1482 pKa = 2.82RR1483 pKa = 11.84LGTYY1487 pKa = 9.8GQGTLGLNIVTSSGVTGFIEE1507 pKa = 4.36AHH1509 pKa = 5.91GEE1511 pKa = 4.13YY1512 pKa = 10.13NDD1514 pKa = 4.11EE1515 pKa = 4.15YY1516 pKa = 11.34SGGGGRR1522 pKa = 11.84AGIRR1526 pKa = 11.84IKK1528 pKa = 10.8FF1529 pKa = 3.53
Molecular weight: 149.96 kDa
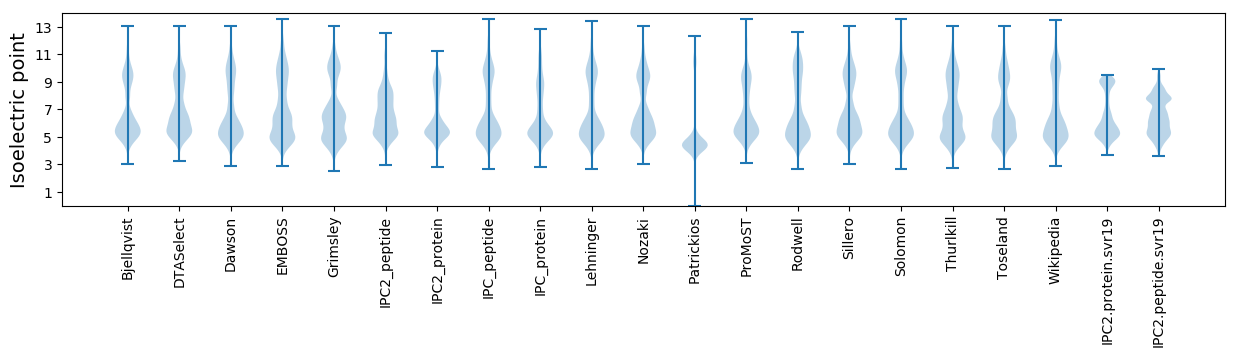
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A318BD78|A0A318BD78_9NOSO Ketosteroid isomerase OS=Nostoc sp. 3335mG OX=2204170 GN=DMC47_20305 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.58GFRR19 pKa = 11.84SRR21 pKa = 11.84MATPGGRR28 pKa = 11.84KK29 pKa = 9.14VIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84AKK41 pKa = 10.7LSAA44 pKa = 4.0
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.58GFRR19 pKa = 11.84SRR21 pKa = 11.84MATPGGRR28 pKa = 11.84KK29 pKa = 9.14VIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84AKK41 pKa = 10.7LSAA44 pKa = 4.0
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2691358 |
25 |
2252 |
311.2 |
33.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.161 ± 0.039 | 0.745 ± 0.007 |
6.056 ± 0.021 | 5.216 ± 0.026 |
3.597 ± 0.016 | 8.943 ± 0.022 |
2.025 ± 0.015 | 5.058 ± 0.018 |
2.813 ± 0.021 | 9.882 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.406 ± 0.013 | 2.492 ± 0.019 |
5.395 ± 0.019 | 3.071 ± 0.013 |
7.42 ± 0.025 | 5.215 ± 0.018 |
5.461 ± 0.025 | 7.29 ± 0.022 |
1.469 ± 0.012 | 2.283 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
