
Vibrio phage VSKK
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; Fibrovirus; Vibrio virus fs1; Vibrio phage VSK
Average proteome isoelectric point is 7.32
Get precalculated fractions of proteins
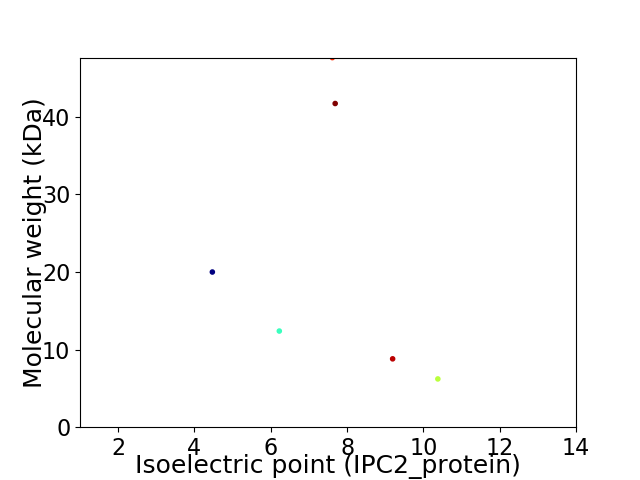
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
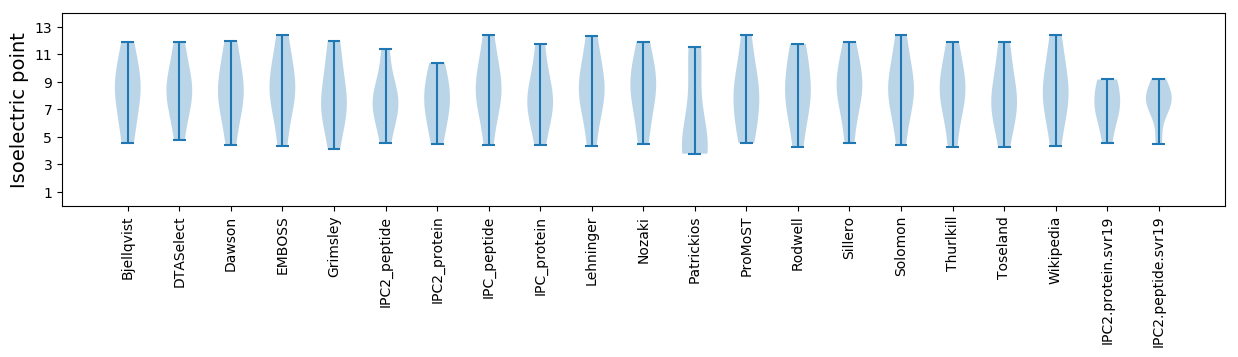
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8W6E1|Q8W6E1_9VIRU Putative major coat protein OS=Vibrio phage VSKK OX=180503 PE=4 SV=1
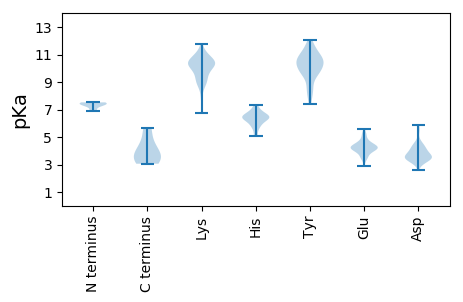
MM1 pKa = 7.29NEE3 pKa = 4.71DD4 pKa = 3.45MNEE7 pKa = 3.73LLTRR11 pKa = 11.84LNSDD15 pKa = 3.45NNKK18 pKa = 9.66QLDD21 pKa = 3.95DD22 pKa = 4.12VNNQLLQLNTQSQRR36 pKa = 11.84IVAQIAKK43 pKa = 9.05QEE45 pKa = 4.11KK46 pKa = 8.44QDD48 pKa = 3.42AAIYY52 pKa = 9.41EE53 pKa = 4.23NTKK56 pKa = 11.01ALIQNLNKK64 pKa = 10.62DD65 pKa = 3.06VTTAVNKK72 pKa = 6.75TTNAVNALGSKK83 pKa = 10.33VDD85 pKa = 3.84GLSDD89 pKa = 3.48AVDD92 pKa = 3.62GLGEE96 pKa = 4.32DD97 pKa = 3.59VSAIKK102 pKa = 10.5DD103 pKa = 3.59AITNVDD109 pKa = 3.12TSGAGISGTCIEE121 pKa = 5.16SDD123 pKa = 3.5TCTGFYY129 pKa = 10.59EE130 pKa = 4.24SGYY133 pKa = 9.37PDD135 pKa = 5.32GISGIFSQHH144 pKa = 5.97FEE146 pKa = 4.26TVSEE150 pKa = 4.52SVHH153 pKa = 6.21DD154 pKa = 3.94TVKK157 pKa = 11.2DD158 pKa = 3.93FMKK161 pKa = 10.12IDD163 pKa = 3.92LSHH166 pKa = 6.56AQRR169 pKa = 11.84PSFSIPVLHH178 pKa = 7.03FGKK181 pKa = 10.26FQLL184 pKa = 4.45
MM1 pKa = 7.29NEE3 pKa = 4.71DD4 pKa = 3.45MNEE7 pKa = 3.73LLTRR11 pKa = 11.84LNSDD15 pKa = 3.45NNKK18 pKa = 9.66QLDD21 pKa = 3.95DD22 pKa = 4.12VNNQLLQLNTQSQRR36 pKa = 11.84IVAQIAKK43 pKa = 9.05QEE45 pKa = 4.11KK46 pKa = 8.44QDD48 pKa = 3.42AAIYY52 pKa = 9.41EE53 pKa = 4.23NTKK56 pKa = 11.01ALIQNLNKK64 pKa = 10.62DD65 pKa = 3.06VTTAVNKK72 pKa = 6.75TTNAVNALGSKK83 pKa = 10.33VDD85 pKa = 3.84GLSDD89 pKa = 3.48AVDD92 pKa = 3.62GLGEE96 pKa = 4.32DD97 pKa = 3.59VSAIKK102 pKa = 10.5DD103 pKa = 3.59AITNVDD109 pKa = 3.12TSGAGISGTCIEE121 pKa = 5.16SDD123 pKa = 3.5TCTGFYY129 pKa = 10.59EE130 pKa = 4.24SGYY133 pKa = 9.37PDD135 pKa = 5.32GISGIFSQHH144 pKa = 5.97FEE146 pKa = 4.26TVSEE150 pKa = 4.52SVHH153 pKa = 6.21DD154 pKa = 3.94TVKK157 pKa = 11.2DD158 pKa = 3.93FMKK161 pKa = 10.12IDD163 pKa = 3.92LSHH166 pKa = 6.56AQRR169 pKa = 11.84PSFSIPVLHH178 pKa = 7.03FGKK181 pKa = 10.26FQLL184 pKa = 4.45
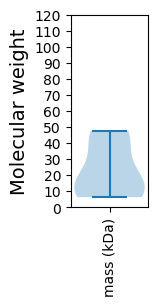
Molecular weight: 19.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8W6D9|Q8W6D9_9VIRU Zot-like protein OS=Vibrio phage VSKK OX=180503 PE=4 SV=1
MM1 pKa = 7.52KK2 pKa = 10.33ADD4 pKa = 5.87LINKK8 pKa = 8.86RR9 pKa = 11.84PKK11 pKa = 10.11RR12 pKa = 11.84YY13 pKa = 8.59STMFLNLVFSKK24 pKa = 10.67KK25 pKa = 10.12LPQAIRR31 pKa = 11.84RR32 pKa = 11.84TGQRR36 pKa = 11.84ASVGQVSRR44 pKa = 11.84RR45 pKa = 11.84EE46 pKa = 3.83WQPRR50 pKa = 11.84RR51 pKa = 11.84DD52 pKa = 3.42
MM1 pKa = 7.52KK2 pKa = 10.33ADD4 pKa = 5.87LINKK8 pKa = 8.86RR9 pKa = 11.84PKK11 pKa = 10.11RR12 pKa = 11.84YY13 pKa = 8.59STMFLNLVFSKK24 pKa = 10.67KK25 pKa = 10.12LPQAIRR31 pKa = 11.84RR32 pKa = 11.84TGQRR36 pKa = 11.84ASVGQVSRR44 pKa = 11.84RR45 pKa = 11.84EE46 pKa = 3.83WQPRR50 pKa = 11.84RR51 pKa = 11.84DD52 pKa = 3.42
Molecular weight: 6.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1205 |
52 |
411 |
200.8 |
22.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.722 ± 1.071 | 1.577 ± 0.366 |
5.809 ± 0.748 | 4.979 ± 0.556 |
4.896 ± 0.302 | 5.809 ± 0.557 |
2.656 ± 0.61 | 7.718 ± 1.182 |
6.722 ± 0.311 | 7.718 ± 0.925 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.905 ± 0.573 | 4.481 ± 0.785 |
4.647 ± 0.567 | 4.398 ± 0.545 |
5.062 ± 1.065 | 6.473 ± 1.053 |
5.809 ± 0.482 | 6.473 ± 0.377 |
1.162 ± 0.314 | 3.983 ± 1.122 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
