
Leptomonas moramango leishbunyavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Leishbuviridae; Shilevirus; Leptomonas shilevirus
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
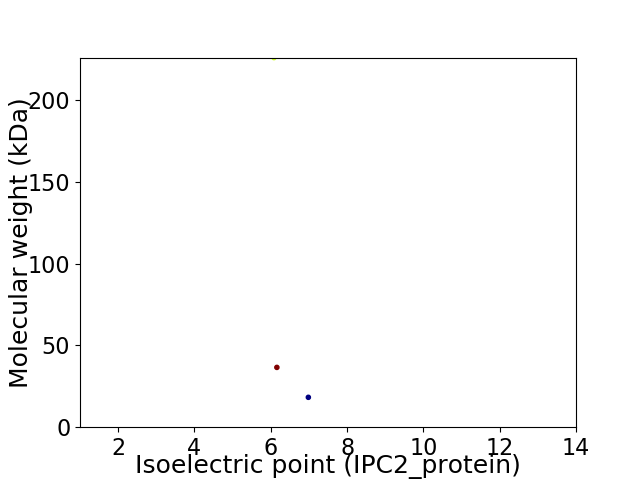
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A191Z3B4|A0A191Z3B4_9VIRU Uncharacterized protein OS=Leptomonas moramango leishbunyavirus OX=1859148 PE=4 SV=1
MM1 pKa = 7.48RR2 pKa = 11.84VSFLSLILLLLLMASGYY19 pKa = 11.2ASDD22 pKa = 5.79DD23 pKa = 4.18PYY25 pKa = 11.72CNTHH29 pKa = 6.13NCLPDD34 pKa = 3.71NEE36 pKa = 4.36PLKK39 pKa = 11.02LVIPSGLVPDD49 pKa = 4.55NYY51 pKa = 11.19LCIFGGTSNFEE62 pKa = 4.13PHH64 pKa = 7.27DD65 pKa = 4.86SIAPEE70 pKa = 3.89QAKK73 pKa = 9.12MEE75 pKa = 4.44YY76 pKa = 9.12YY77 pKa = 10.47AKK79 pKa = 10.17HH80 pKa = 6.43DD81 pKa = 4.03AYY83 pKa = 9.96ICRR86 pKa = 11.84NTPNVLEE93 pKa = 5.17ASYY96 pKa = 10.12KK97 pKa = 9.65WYY99 pKa = 9.9MEE101 pKa = 4.08AGKK104 pKa = 10.06NLVTVLVDD112 pKa = 3.23VDD114 pKa = 4.33RR115 pKa = 11.84SSSPVVPPTSQIRR128 pKa = 11.84ASSNYY133 pKa = 10.33LIDD136 pKa = 3.32AHH138 pKa = 8.06FEE140 pKa = 3.8MGMLNIRR147 pKa = 11.84EE148 pKa = 4.24PYY150 pKa = 8.52QRR152 pKa = 11.84CRR154 pKa = 11.84PGPSDD159 pKa = 3.77WIPRR163 pKa = 11.84RR164 pKa = 11.84KK165 pKa = 9.79SEE167 pKa = 4.41CIHH170 pKa = 6.0RR171 pKa = 11.84QSSYY175 pKa = 10.31NIGMAVVYY183 pKa = 9.78PSEE186 pKa = 3.71ILQNYY191 pKa = 9.44KK192 pKa = 10.07IDD194 pKa = 3.64VDD196 pKa = 3.73PDD198 pKa = 3.65GVVISTSKK206 pKa = 11.02GDD208 pKa = 2.9WSTAIRR214 pKa = 11.84AEE216 pKa = 4.01CANYY220 pKa = 9.97KK221 pKa = 9.7GLRR224 pKa = 11.84RR225 pKa = 11.84HH226 pKa = 7.16LIGTTLRR233 pKa = 11.84IRR235 pKa = 11.84SKK237 pKa = 11.32DD238 pKa = 3.39SLACLITGSRR248 pKa = 11.84QTLYY252 pKa = 11.07KK253 pKa = 10.38DD254 pKa = 2.54ITMRR258 pKa = 11.84YY259 pKa = 8.91AFEE262 pKa = 4.71SKK264 pKa = 9.3TGCEE268 pKa = 4.13TKK270 pKa = 10.77LKK272 pKa = 10.56ARR274 pKa = 11.84QLFDD278 pKa = 2.9SWYY281 pKa = 9.06SPSFFLSVGPVDD293 pKa = 4.35RR294 pKa = 11.84LPVLYY299 pKa = 9.83EE300 pKa = 4.02SCKK303 pKa = 10.75DD304 pKa = 3.65NQDD307 pKa = 3.46GEE309 pKa = 4.47FFRR312 pKa = 11.84DD313 pKa = 4.29DD314 pKa = 3.93YY315 pKa = 11.47KK316 pKa = 10.32QQCYY320 pKa = 10.46ASS322 pKa = 3.61
MM1 pKa = 7.48RR2 pKa = 11.84VSFLSLILLLLLMASGYY19 pKa = 11.2ASDD22 pKa = 5.79DD23 pKa = 4.18PYY25 pKa = 11.72CNTHH29 pKa = 6.13NCLPDD34 pKa = 3.71NEE36 pKa = 4.36PLKK39 pKa = 11.02LVIPSGLVPDD49 pKa = 4.55NYY51 pKa = 11.19LCIFGGTSNFEE62 pKa = 4.13PHH64 pKa = 7.27DD65 pKa = 4.86SIAPEE70 pKa = 3.89QAKK73 pKa = 9.12MEE75 pKa = 4.44YY76 pKa = 9.12YY77 pKa = 10.47AKK79 pKa = 10.17HH80 pKa = 6.43DD81 pKa = 4.03AYY83 pKa = 9.96ICRR86 pKa = 11.84NTPNVLEE93 pKa = 5.17ASYY96 pKa = 10.12KK97 pKa = 9.65WYY99 pKa = 9.9MEE101 pKa = 4.08AGKK104 pKa = 10.06NLVTVLVDD112 pKa = 3.23VDD114 pKa = 4.33RR115 pKa = 11.84SSSPVVPPTSQIRR128 pKa = 11.84ASSNYY133 pKa = 10.33LIDD136 pKa = 3.32AHH138 pKa = 8.06FEE140 pKa = 3.8MGMLNIRR147 pKa = 11.84EE148 pKa = 4.24PYY150 pKa = 8.52QRR152 pKa = 11.84CRR154 pKa = 11.84PGPSDD159 pKa = 3.77WIPRR163 pKa = 11.84RR164 pKa = 11.84KK165 pKa = 9.79SEE167 pKa = 4.41CIHH170 pKa = 6.0RR171 pKa = 11.84QSSYY175 pKa = 10.31NIGMAVVYY183 pKa = 9.78PSEE186 pKa = 3.71ILQNYY191 pKa = 9.44KK192 pKa = 10.07IDD194 pKa = 3.64VDD196 pKa = 3.73PDD198 pKa = 3.65GVVISTSKK206 pKa = 11.02GDD208 pKa = 2.9WSTAIRR214 pKa = 11.84AEE216 pKa = 4.01CANYY220 pKa = 9.97KK221 pKa = 9.7GLRR224 pKa = 11.84RR225 pKa = 11.84HH226 pKa = 7.16LIGTTLRR233 pKa = 11.84IRR235 pKa = 11.84SKK237 pKa = 11.32DD238 pKa = 3.39SLACLITGSRR248 pKa = 11.84QTLYY252 pKa = 11.07KK253 pKa = 10.38DD254 pKa = 2.54ITMRR258 pKa = 11.84YY259 pKa = 8.91AFEE262 pKa = 4.71SKK264 pKa = 9.3TGCEE268 pKa = 4.13TKK270 pKa = 10.77LKK272 pKa = 10.56ARR274 pKa = 11.84QLFDD278 pKa = 2.9SWYY281 pKa = 9.06SPSFFLSVGPVDD293 pKa = 4.35RR294 pKa = 11.84LPVLYY299 pKa = 9.83EE300 pKa = 4.02SCKK303 pKa = 10.75DD304 pKa = 3.65NQDD307 pKa = 3.46GEE309 pKa = 4.47FFRR312 pKa = 11.84DD313 pKa = 4.29DD314 pKa = 3.93YY315 pKa = 11.47KK316 pKa = 10.32QQCYY320 pKa = 10.46ASS322 pKa = 3.61
Molecular weight: 36.58 kDa
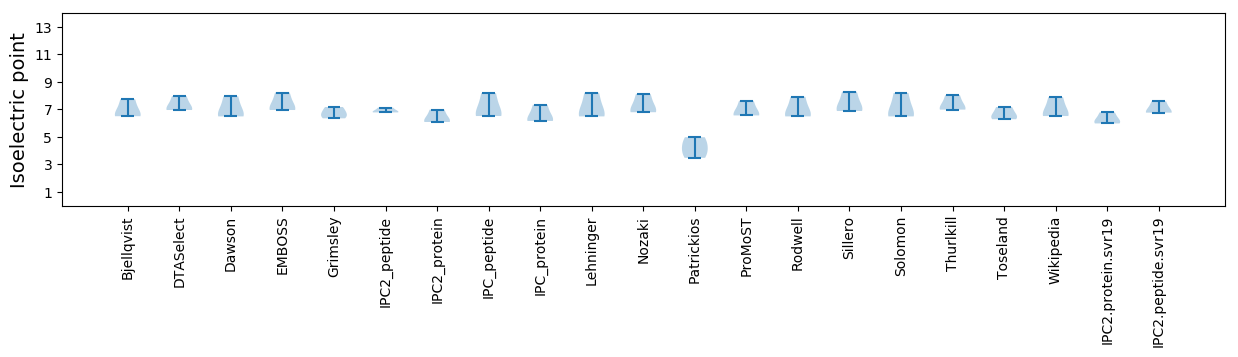
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A191Z3B4|A0A191Z3B4_9VIRU Uncharacterized protein OS=Leptomonas moramango leishbunyavirus OX=1859148 PE=4 SV=1
MM1 pKa = 7.41TSVFLPFDD9 pKa = 4.16LEE11 pKa = 4.1LTNALEE17 pKa = 4.4YY18 pKa = 10.82KK19 pKa = 10.26GVSPFEE25 pKa = 4.01TRR27 pKa = 11.84TKK29 pKa = 10.08IVAAGKK35 pKa = 9.19EE36 pKa = 4.19NQGKK40 pKa = 9.07LVALMVACRR49 pKa = 11.84GTNIFKK55 pKa = 10.49IVTKK59 pKa = 10.71AKK61 pKa = 10.66DD62 pKa = 3.26PALSTKK68 pKa = 9.79IVAISEE74 pKa = 4.04EE75 pKa = 4.25LSRR78 pKa = 11.84EE79 pKa = 4.05FAVSLAHH86 pKa = 6.88IASAFPEE93 pKa = 4.41VVYY96 pKa = 8.57DD97 pKa = 3.57TRR99 pKa = 11.84IKK101 pKa = 10.0ISSSISVNTLAFLKK115 pKa = 10.85HH116 pKa = 6.3NGLTHH121 pKa = 6.58EE122 pKa = 4.46KK123 pKa = 9.24WLLANEE129 pKa = 4.43EE130 pKa = 4.06FCKK133 pKa = 10.73LVGLDD138 pKa = 3.54FAKK141 pKa = 10.57FLAVEE146 pKa = 3.96EE147 pKa = 4.99VIWKK151 pKa = 9.97DD152 pKa = 3.28DD153 pKa = 3.65SFEE156 pKa = 5.03KK157 pKa = 10.51IVGARR162 pKa = 11.84KK163 pKa = 8.94PLL165 pKa = 3.58
MM1 pKa = 7.41TSVFLPFDD9 pKa = 4.16LEE11 pKa = 4.1LTNALEE17 pKa = 4.4YY18 pKa = 10.82KK19 pKa = 10.26GVSPFEE25 pKa = 4.01TRR27 pKa = 11.84TKK29 pKa = 10.08IVAAGKK35 pKa = 9.19EE36 pKa = 4.19NQGKK40 pKa = 9.07LVALMVACRR49 pKa = 11.84GTNIFKK55 pKa = 10.49IVTKK59 pKa = 10.71AKK61 pKa = 10.66DD62 pKa = 3.26PALSTKK68 pKa = 9.79IVAISEE74 pKa = 4.04EE75 pKa = 4.25LSRR78 pKa = 11.84EE79 pKa = 4.05FAVSLAHH86 pKa = 6.88IASAFPEE93 pKa = 4.41VVYY96 pKa = 8.57DD97 pKa = 3.57TRR99 pKa = 11.84IKK101 pKa = 10.0ISSSISVNTLAFLKK115 pKa = 10.85HH116 pKa = 6.3NGLTHH121 pKa = 6.58EE122 pKa = 4.46KK123 pKa = 9.24WLLANEE129 pKa = 4.43EE130 pKa = 4.06FCKK133 pKa = 10.73LVGLDD138 pKa = 3.54FAKK141 pKa = 10.57FLAVEE146 pKa = 3.96EE147 pKa = 4.99VIWKK151 pKa = 9.97DD152 pKa = 3.28DD153 pKa = 3.65SFEE156 pKa = 5.03KK157 pKa = 10.51IVGARR162 pKa = 11.84KK163 pKa = 8.94PLL165 pKa = 3.58
Molecular weight: 18.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2466 |
165 |
1979 |
822.0 |
93.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.799 ± 1.044 | 1.906 ± 0.406 |
5.231 ± 0.452 | 6.529 ± 0.536 |
4.704 ± 0.488 | 4.461 ± 0.137 |
2.96 ± 0.519 | 6.772 ± 0.277 |
6.042 ± 0.771 | 9.976 ± 0.271 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.149 ± 0.197 | 4.623 ± 0.284 |
4.582 ± 0.455 | 3.487 ± 0.748 |
5.069 ± 0.443 | 9.611 ± 0.517 |
5.353 ± 0.269 | 5.474 ± 0.878 |
1.703 ± 0.22 | 3.569 ± 0.727 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
