
Lymantria dispar cypovirus 14
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Cypovirus; Cypovirus 14
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
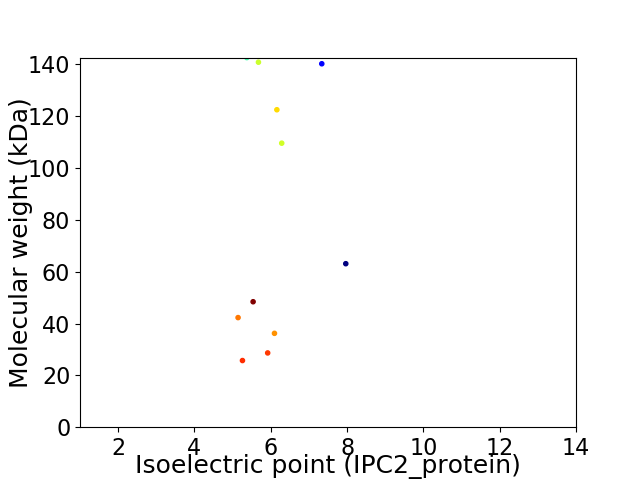
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
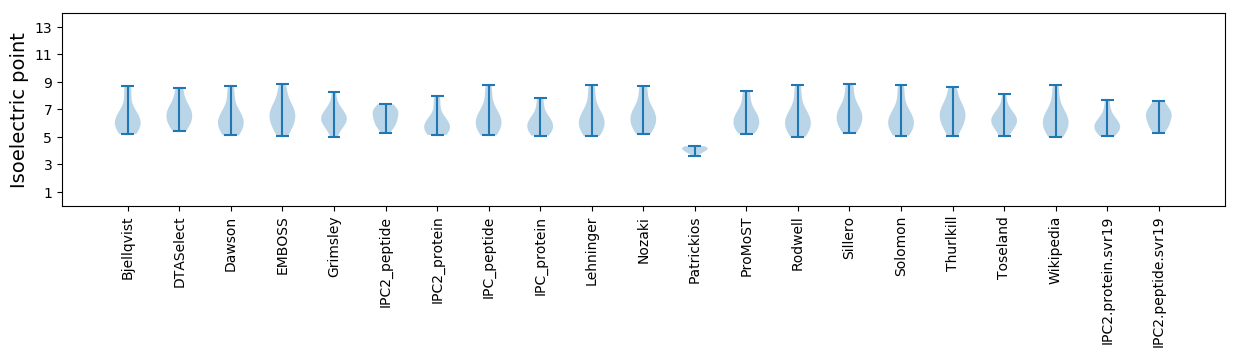
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q91IE6|Q91IE6_9REOV Uncharacterized protein OS=Lymantria dispar cypovirus 14 OX=165429 PE=4 SV=1
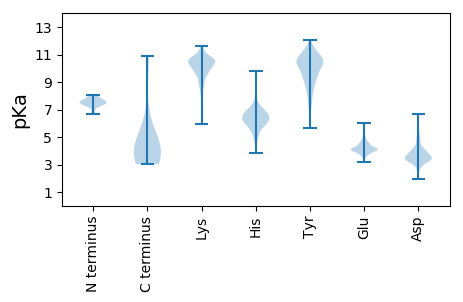
MM1 pKa = 7.47SISRR5 pKa = 11.84SEE7 pKa = 3.68ITDD10 pKa = 2.8IGGRR14 pKa = 11.84LLYY17 pKa = 9.24TVYY20 pKa = 9.53YY21 pKa = 6.85TTLDD25 pKa = 3.23NYY27 pKa = 11.37KK28 pKa = 10.35LIKK31 pKa = 10.72NEE33 pKa = 3.85FTSNGLVHH41 pKa = 6.85KK42 pKa = 10.23AVNSKK47 pKa = 10.84ALTNVRR53 pKa = 11.84MLIEE57 pKa = 4.61NFTKK61 pKa = 10.66KK62 pKa = 10.34NINIIYY68 pKa = 9.63IEE70 pKa = 4.05QGSILASNITTLALTYY86 pKa = 11.09KK87 pKa = 9.24MLIVIQSPKK96 pKa = 10.07EE97 pKa = 3.75KK98 pKa = 9.84TIAGAIKK105 pKa = 8.88WRR107 pKa = 11.84NAVGSRR113 pKa = 11.84INGPVHH119 pKa = 6.08MALSDD124 pKa = 3.9YY125 pKa = 10.7IALIKK130 pKa = 10.54AEE132 pKa = 4.18RR133 pKa = 11.84EE134 pKa = 3.92KK135 pKa = 10.91AVQSSTVEE143 pKa = 4.0PVSVVPVAKK152 pKa = 10.55LMQEE156 pKa = 4.07PVHH159 pKa = 8.01VDD161 pKa = 3.0VTQAEE166 pKa = 4.24VHH168 pKa = 5.78IQNEE172 pKa = 4.47EE173 pKa = 4.12VKK175 pKa = 10.45VDD177 pKa = 3.25HH178 pKa = 6.65SVNEE182 pKa = 3.97EE183 pKa = 4.07SAPSDD188 pKa = 3.18SVTEE192 pKa = 3.97VKK194 pKa = 9.97QQNVCSTCDD203 pKa = 3.33KK204 pKa = 10.66PIAEE208 pKa = 4.6YY209 pKa = 11.25ACMRR213 pKa = 11.84DD214 pKa = 5.72DD215 pKa = 5.17NDD217 pKa = 4.18CSCDD221 pKa = 4.12AMDD224 pKa = 5.57DD225 pKa = 3.84ALNKK229 pKa = 10.61SFDD232 pKa = 4.21TIDD235 pKa = 3.43SDD237 pKa = 5.15GATLDD242 pKa = 4.07LPALSMRR249 pKa = 11.84RR250 pKa = 11.84VYY252 pKa = 11.14DD253 pKa = 3.69SVCIGCADD261 pKa = 3.67APLEE265 pKa = 3.78QLVLHH270 pKa = 6.76IEE272 pKa = 4.1LSEE275 pKa = 4.0LMTRR279 pKa = 11.84ARR281 pKa = 11.84LLLSKK286 pKa = 10.23FMKK289 pKa = 10.4VYY291 pKa = 10.65LVPSDD296 pKa = 4.82LDD298 pKa = 3.64WNEE301 pKa = 3.96LRR303 pKa = 11.84TITIDD308 pKa = 4.99GITLQYY314 pKa = 9.64RR315 pKa = 11.84TTLKK319 pKa = 10.68DD320 pKa = 3.5SNSILCEE327 pKa = 3.78SEE329 pKa = 5.3LISKK333 pKa = 8.9VFEE336 pKa = 4.05MYY338 pKa = 10.27RR339 pKa = 11.84GSEE342 pKa = 3.87RR343 pKa = 11.84LFQYY347 pKa = 10.83KK348 pKa = 10.24DD349 pKa = 3.42QLCSMIGMHH358 pKa = 6.83ISGDD362 pKa = 3.59PTIFICSNACGLMLAKK378 pKa = 9.55MM379 pKa = 4.32
MM1 pKa = 7.47SISRR5 pKa = 11.84SEE7 pKa = 3.68ITDD10 pKa = 2.8IGGRR14 pKa = 11.84LLYY17 pKa = 9.24TVYY20 pKa = 9.53YY21 pKa = 6.85TTLDD25 pKa = 3.23NYY27 pKa = 11.37KK28 pKa = 10.35LIKK31 pKa = 10.72NEE33 pKa = 3.85FTSNGLVHH41 pKa = 6.85KK42 pKa = 10.23AVNSKK47 pKa = 10.84ALTNVRR53 pKa = 11.84MLIEE57 pKa = 4.61NFTKK61 pKa = 10.66KK62 pKa = 10.34NINIIYY68 pKa = 9.63IEE70 pKa = 4.05QGSILASNITTLALTYY86 pKa = 11.09KK87 pKa = 9.24MLIVIQSPKK96 pKa = 10.07EE97 pKa = 3.75KK98 pKa = 9.84TIAGAIKK105 pKa = 8.88WRR107 pKa = 11.84NAVGSRR113 pKa = 11.84INGPVHH119 pKa = 6.08MALSDD124 pKa = 3.9YY125 pKa = 10.7IALIKK130 pKa = 10.54AEE132 pKa = 4.18RR133 pKa = 11.84EE134 pKa = 3.92KK135 pKa = 10.91AVQSSTVEE143 pKa = 4.0PVSVVPVAKK152 pKa = 10.55LMQEE156 pKa = 4.07PVHH159 pKa = 8.01VDD161 pKa = 3.0VTQAEE166 pKa = 4.24VHH168 pKa = 5.78IQNEE172 pKa = 4.47EE173 pKa = 4.12VKK175 pKa = 10.45VDD177 pKa = 3.25HH178 pKa = 6.65SVNEE182 pKa = 3.97EE183 pKa = 4.07SAPSDD188 pKa = 3.18SVTEE192 pKa = 3.97VKK194 pKa = 9.97QQNVCSTCDD203 pKa = 3.33KK204 pKa = 10.66PIAEE208 pKa = 4.6YY209 pKa = 11.25ACMRR213 pKa = 11.84DD214 pKa = 5.72DD215 pKa = 5.17NDD217 pKa = 4.18CSCDD221 pKa = 4.12AMDD224 pKa = 5.57DD225 pKa = 3.84ALNKK229 pKa = 10.61SFDD232 pKa = 4.21TIDD235 pKa = 3.43SDD237 pKa = 5.15GATLDD242 pKa = 4.07LPALSMRR249 pKa = 11.84RR250 pKa = 11.84VYY252 pKa = 11.14DD253 pKa = 3.69SVCIGCADD261 pKa = 3.67APLEE265 pKa = 3.78QLVLHH270 pKa = 6.76IEE272 pKa = 4.1LSEE275 pKa = 4.0LMTRR279 pKa = 11.84ARR281 pKa = 11.84LLLSKK286 pKa = 10.23FMKK289 pKa = 10.4VYY291 pKa = 10.65LVPSDD296 pKa = 4.82LDD298 pKa = 3.64WNEE301 pKa = 3.96LRR303 pKa = 11.84TITIDD308 pKa = 4.99GITLQYY314 pKa = 9.64RR315 pKa = 11.84TTLKK319 pKa = 10.68DD320 pKa = 3.5SNSILCEE327 pKa = 3.78SEE329 pKa = 5.3LISKK333 pKa = 8.9VFEE336 pKa = 4.05MYY338 pKa = 10.27RR339 pKa = 11.84GSEE342 pKa = 3.87RR343 pKa = 11.84LFQYY347 pKa = 10.83KK348 pKa = 10.24DD349 pKa = 3.42QLCSMIGMHH358 pKa = 6.83ISGDD362 pKa = 3.59PTIFICSNACGLMLAKK378 pKa = 9.55MM379 pKa = 4.32
Molecular weight: 42.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q91IE8|Q91IE8_9REOV Uncharacterized protein OS=Lymantria dispar cypovirus 14 OX=165429 PE=4 SV=1
MM1 pKa = 7.5LAIDD5 pKa = 3.74YY6 pKa = 8.23LQNEE10 pKa = 4.73RR11 pKa = 11.84LYY13 pKa = 9.12HH14 pKa = 5.57TYY16 pKa = 9.5MLKK19 pKa = 9.55TKK21 pKa = 9.97TNEE24 pKa = 3.83NNNAPTPEE32 pKa = 3.97VKK34 pKa = 10.28QFLLQSSIAHH44 pKa = 6.46KK45 pKa = 10.52LLRR48 pKa = 11.84AYY50 pKa = 10.69SNKK53 pKa = 8.92TKK55 pKa = 10.8GVIKK59 pKa = 10.63DD60 pKa = 3.55PALLGAIISTGSYY73 pKa = 10.52DD74 pKa = 3.54VQVSNKK80 pKa = 8.98KK81 pKa = 9.7DD82 pKa = 3.27RR83 pKa = 11.84KK84 pKa = 9.72IVGKK88 pKa = 10.31AIDD91 pKa = 3.92GANVKK96 pKa = 8.46VTVKK100 pKa = 10.4KK101 pKa = 10.44GRR103 pKa = 11.84KK104 pKa = 8.83KK105 pKa = 10.19RR106 pKa = 11.84DD107 pKa = 3.04VMADD111 pKa = 3.37MPAQTSLAADD121 pKa = 3.46QEE123 pKa = 4.91GVLTLEE129 pKa = 4.96DD130 pKa = 3.9MNEE133 pKa = 3.83LSEE136 pKa = 4.18MAKK139 pKa = 10.38KK140 pKa = 10.45LRR142 pKa = 11.84TVYY145 pKa = 9.07NTSEE149 pKa = 4.45SKK151 pKa = 10.74SLWYY155 pKa = 10.04IKK157 pKa = 9.67DD158 pKa = 3.19EE159 pKa = 4.51RR160 pKa = 11.84YY161 pKa = 11.22SMLIPSYY168 pKa = 9.14ITAIMPFMPRR178 pKa = 11.84RR179 pKa = 11.84EE180 pKa = 4.04IEE182 pKa = 3.93LSEE185 pKa = 4.38KK186 pKa = 10.61GRR188 pKa = 11.84MVVPLDD194 pKa = 3.55LKK196 pKa = 11.03HH197 pKa = 6.12IFRR200 pKa = 11.84NIVVVRR206 pKa = 11.84EE207 pKa = 4.0YY208 pKa = 11.47NEE210 pKa = 3.71NQLRR214 pKa = 11.84NNKK217 pKa = 8.74PCNSLLINASVLSDD231 pKa = 3.38SVYY234 pKa = 9.1MQKK237 pKa = 10.67GYY239 pKa = 9.86QVFTNSVIVYY249 pKa = 5.91TASWRR254 pKa = 11.84NRR256 pKa = 11.84EE257 pKa = 3.4VDD259 pKa = 5.27FIMFDD264 pKa = 3.28TRR266 pKa = 11.84NVVDD270 pKa = 5.6NIDD273 pKa = 3.79GSDD276 pKa = 3.63VEE278 pKa = 4.93IVWKK282 pKa = 10.27EE283 pKa = 3.63LIKK286 pKa = 10.6YY287 pKa = 7.81ARR289 pKa = 11.84PVANLYY295 pKa = 7.7EE296 pKa = 4.26TLEE299 pKa = 4.19PEE301 pKa = 4.1MVIKK305 pKa = 10.76KK306 pKa = 9.87KK307 pKa = 10.87GPTIYY312 pKa = 10.49QPIRR316 pKa = 11.84GATYY320 pKa = 10.45AVLLKK325 pKa = 10.57FRR327 pKa = 11.84EE328 pKa = 4.13QCLSKK333 pKa = 9.33TQLKK337 pKa = 10.72VLMTFIGNKK346 pKa = 9.92GFGEE350 pKa = 4.41TSFLRR355 pKa = 11.84DD356 pKa = 3.31VIAALNSKK364 pKa = 10.45LGDD367 pKa = 3.61NAAGAIDD374 pKa = 4.06SDD376 pKa = 4.02WYY378 pKa = 10.88GIWEE382 pKa = 4.24YY383 pKa = 11.26KK384 pKa = 9.08RR385 pKa = 11.84TVEE388 pKa = 4.97GIEE391 pKa = 4.6LPLEE395 pKa = 4.08YY396 pKa = 10.18NVCVEE401 pKa = 4.13QLEE404 pKa = 4.53SGPSIFEE411 pKa = 4.21YY412 pKa = 10.61YY413 pKa = 10.72AEE415 pKa = 4.69QILSAANIKK424 pKa = 9.62TSEE427 pKa = 4.18QYY429 pKa = 10.83FKK431 pKa = 10.99TKK433 pKa = 9.43IQQRR437 pKa = 11.84EE438 pKa = 3.98TMINEE443 pKa = 4.35CKK445 pKa = 10.62SKK447 pKa = 11.18VEE449 pKa = 3.89QHH451 pKa = 5.61WLNGKK456 pKa = 8.47QSPEE460 pKa = 3.6YY461 pKa = 9.61QFNVRR466 pKa = 11.84VTTSKK471 pKa = 8.94TAPRR475 pKa = 11.84ILILNQHH482 pKa = 5.98TVTQDD487 pKa = 3.11VTAGRR492 pKa = 11.84SDD494 pKa = 2.98INLILKK500 pKa = 8.78PIIDD504 pKa = 4.35PVSMLLLRR512 pKa = 11.84DD513 pKa = 3.16RR514 pKa = 11.84AMPAEE519 pKa = 4.07LLLYY523 pKa = 7.34EE524 pKa = 4.39TYY526 pKa = 11.57GMLTSYY532 pKa = 10.61VHH534 pKa = 6.13TSMYY538 pKa = 10.61ACEE541 pKa = 4.11LLRR544 pKa = 11.84FIEE547 pKa = 4.3YY548 pKa = 10.17DD549 pKa = 2.94WW550 pKa = 5.68
MM1 pKa = 7.5LAIDD5 pKa = 3.74YY6 pKa = 8.23LQNEE10 pKa = 4.73RR11 pKa = 11.84LYY13 pKa = 9.12HH14 pKa = 5.57TYY16 pKa = 9.5MLKK19 pKa = 9.55TKK21 pKa = 9.97TNEE24 pKa = 3.83NNNAPTPEE32 pKa = 3.97VKK34 pKa = 10.28QFLLQSSIAHH44 pKa = 6.46KK45 pKa = 10.52LLRR48 pKa = 11.84AYY50 pKa = 10.69SNKK53 pKa = 8.92TKK55 pKa = 10.8GVIKK59 pKa = 10.63DD60 pKa = 3.55PALLGAIISTGSYY73 pKa = 10.52DD74 pKa = 3.54VQVSNKK80 pKa = 8.98KK81 pKa = 9.7DD82 pKa = 3.27RR83 pKa = 11.84KK84 pKa = 9.72IVGKK88 pKa = 10.31AIDD91 pKa = 3.92GANVKK96 pKa = 8.46VTVKK100 pKa = 10.4KK101 pKa = 10.44GRR103 pKa = 11.84KK104 pKa = 8.83KK105 pKa = 10.19RR106 pKa = 11.84DD107 pKa = 3.04VMADD111 pKa = 3.37MPAQTSLAADD121 pKa = 3.46QEE123 pKa = 4.91GVLTLEE129 pKa = 4.96DD130 pKa = 3.9MNEE133 pKa = 3.83LSEE136 pKa = 4.18MAKK139 pKa = 10.38KK140 pKa = 10.45LRR142 pKa = 11.84TVYY145 pKa = 9.07NTSEE149 pKa = 4.45SKK151 pKa = 10.74SLWYY155 pKa = 10.04IKK157 pKa = 9.67DD158 pKa = 3.19EE159 pKa = 4.51RR160 pKa = 11.84YY161 pKa = 11.22SMLIPSYY168 pKa = 9.14ITAIMPFMPRR178 pKa = 11.84RR179 pKa = 11.84EE180 pKa = 4.04IEE182 pKa = 3.93LSEE185 pKa = 4.38KK186 pKa = 10.61GRR188 pKa = 11.84MVVPLDD194 pKa = 3.55LKK196 pKa = 11.03HH197 pKa = 6.12IFRR200 pKa = 11.84NIVVVRR206 pKa = 11.84EE207 pKa = 4.0YY208 pKa = 11.47NEE210 pKa = 3.71NQLRR214 pKa = 11.84NNKK217 pKa = 8.74PCNSLLINASVLSDD231 pKa = 3.38SVYY234 pKa = 9.1MQKK237 pKa = 10.67GYY239 pKa = 9.86QVFTNSVIVYY249 pKa = 5.91TASWRR254 pKa = 11.84NRR256 pKa = 11.84EE257 pKa = 3.4VDD259 pKa = 5.27FIMFDD264 pKa = 3.28TRR266 pKa = 11.84NVVDD270 pKa = 5.6NIDD273 pKa = 3.79GSDD276 pKa = 3.63VEE278 pKa = 4.93IVWKK282 pKa = 10.27EE283 pKa = 3.63LIKK286 pKa = 10.6YY287 pKa = 7.81ARR289 pKa = 11.84PVANLYY295 pKa = 7.7EE296 pKa = 4.26TLEE299 pKa = 4.19PEE301 pKa = 4.1MVIKK305 pKa = 10.76KK306 pKa = 9.87KK307 pKa = 10.87GPTIYY312 pKa = 10.49QPIRR316 pKa = 11.84GATYY320 pKa = 10.45AVLLKK325 pKa = 10.57FRR327 pKa = 11.84EE328 pKa = 4.13QCLSKK333 pKa = 9.33TQLKK337 pKa = 10.72VLMTFIGNKK346 pKa = 9.92GFGEE350 pKa = 4.41TSFLRR355 pKa = 11.84DD356 pKa = 3.31VIAALNSKK364 pKa = 10.45LGDD367 pKa = 3.61NAAGAIDD374 pKa = 4.06SDD376 pKa = 4.02WYY378 pKa = 10.88GIWEE382 pKa = 4.24YY383 pKa = 11.26KK384 pKa = 9.08RR385 pKa = 11.84TVEE388 pKa = 4.97GIEE391 pKa = 4.6LPLEE395 pKa = 4.08YY396 pKa = 10.18NVCVEE401 pKa = 4.13QLEE404 pKa = 4.53SGPSIFEE411 pKa = 4.21YY412 pKa = 10.61YY413 pKa = 10.72AEE415 pKa = 4.69QILSAANIKK424 pKa = 9.62TSEE427 pKa = 4.18QYY429 pKa = 10.83FKK431 pKa = 10.99TKK433 pKa = 9.43IQQRR437 pKa = 11.84EE438 pKa = 3.98TMINEE443 pKa = 4.35CKK445 pKa = 10.62SKK447 pKa = 11.18VEE449 pKa = 3.89QHH451 pKa = 5.61WLNGKK456 pKa = 8.47QSPEE460 pKa = 3.6YY461 pKa = 9.61QFNVRR466 pKa = 11.84VTTSKK471 pKa = 8.94TAPRR475 pKa = 11.84ILILNQHH482 pKa = 5.98TVTQDD487 pKa = 3.11VTAGRR492 pKa = 11.84SDD494 pKa = 2.98INLILKK500 pKa = 8.78PIIDD504 pKa = 4.35PVSMLLLRR512 pKa = 11.84DD513 pKa = 3.16RR514 pKa = 11.84AMPAEE519 pKa = 4.07LLLYY523 pKa = 7.34EE524 pKa = 4.39TYY526 pKa = 11.57GMLTSYY532 pKa = 10.61VHH534 pKa = 6.13TSMYY538 pKa = 10.61ACEE541 pKa = 4.11LLRR544 pKa = 11.84FIEE547 pKa = 4.3YY548 pKa = 10.17DD549 pKa = 2.94WW550 pKa = 5.68
Molecular weight: 63.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7925 |
233 |
1267 |
720.5 |
81.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.562 ± 0.234 | 1.186 ± 0.323 |
6.019 ± 0.232 | 5.716 ± 0.255 |
3.899 ± 0.214 | 4.808 ± 0.27 |
2.259 ± 0.17 | 6.524 ± 0.407 |
4.656 ± 0.572 | 9.274 ± 0.413 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.331 ± 0.165 | 6.057 ± 0.456 |
3.836 ± 0.313 | 3.95 ± 0.321 |
6.145 ± 0.3 | 7.155 ± 0.217 |
6.448 ± 0.332 | 7.066 ± 0.561 |
0.782 ± 0.065 | 4.328 ± 0.261 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
