
McMurdo Ice Shelf pond-associated circular DNA virus-2
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.73
Get precalculated fractions of proteins
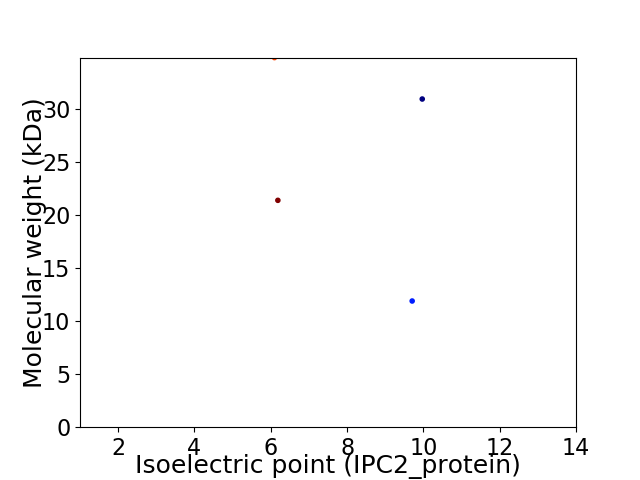
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075LZ73|A0A075LZ73_9VIRU Uncharacterized protein OS=McMurdo Ice Shelf pond-associated circular DNA virus-2 OX=1521386 PE=4 SV=1
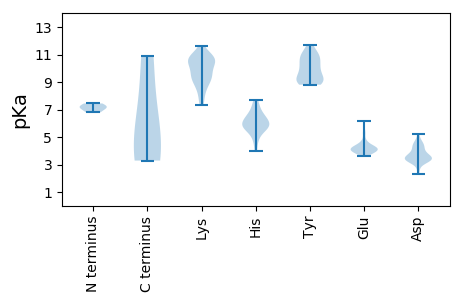
MM1 pKa = 7.15VNARR5 pKa = 11.84CVCVTIHH12 pKa = 6.43VDD14 pKa = 3.58NIFWEE19 pKa = 4.34LQKK22 pKa = 10.47WNQSLTYY29 pKa = 10.54GIGQLEE35 pKa = 4.4LGLNGSTHH43 pKa = 3.99WQMYY47 pKa = 9.07FEE49 pKa = 5.24NNTAISLTQWKK60 pKa = 9.17QLLGCKK66 pKa = 9.29RR67 pKa = 11.84AHH69 pKa = 5.68VEE71 pKa = 4.13TRR73 pKa = 11.84KK74 pKa = 8.84GTALLAIEE82 pKa = 4.28YY83 pKa = 9.15CKK85 pKa = 10.71KK86 pKa = 10.65EE87 pKa = 3.99EE88 pKa = 4.19TRR90 pKa = 11.84LHH92 pKa = 6.14GPGTSFEE99 pKa = 4.27FGKK102 pKa = 7.69TTEE105 pKa = 4.11EE106 pKa = 4.04SVKK109 pKa = 10.43RR110 pKa = 11.84KK111 pKa = 10.51SPDD114 pKa = 3.14SPYY117 pKa = 11.16KK118 pKa = 10.33KK119 pKa = 10.42ALNCLSYY126 pKa = 10.95ADD128 pKa = 4.74SIAIIKK134 pKa = 10.46DD135 pKa = 3.32EE136 pKa = 4.17APRR139 pKa = 11.84DD140 pKa = 3.4YY141 pKa = 10.94VIYY144 pKa = 10.02HH145 pKa = 5.7NQVTSTLKK153 pKa = 10.9AIFKK157 pKa = 8.87PVWNKK162 pKa = 9.28TPGLVFNLEE171 pKa = 4.18PFPEE175 pKa = 4.25EE176 pKa = 3.98TLNTRR181 pKa = 11.84AVIFMGRR188 pKa = 11.84SGLGKK193 pKa = 7.31TQFAISHH200 pKa = 6.41FKK202 pKa = 10.85DD203 pKa = 3.39PLIVSHH209 pKa = 7.66IDD211 pKa = 3.39DD212 pKa = 4.6LKK214 pKa = 11.21KK215 pKa = 10.88LNPTIDD221 pKa = 4.37GIIFDD226 pKa = 5.22DD227 pKa = 4.39MNFQHH232 pKa = 7.06WPPTACIHH240 pKa = 5.92LCDD243 pKa = 3.88MEE245 pKa = 4.71LPRR248 pKa = 11.84SINVKK253 pKa = 9.84YY254 pKa = 9.03GTVEE258 pKa = 3.79IPANTPRR265 pKa = 11.84IFTSNRR271 pKa = 11.84DD272 pKa = 3.59FEE274 pKa = 5.28EE275 pKa = 4.15IWSKK279 pKa = 11.59DD280 pKa = 3.32CTFEE284 pKa = 3.75EE285 pKa = 4.43MNAIRR290 pKa = 11.84RR291 pKa = 11.84RR292 pKa = 11.84CDD294 pKa = 2.33ITTFDD299 pKa = 3.72NEE301 pKa = 4.33LYY303 pKa = 10.88
MM1 pKa = 7.15VNARR5 pKa = 11.84CVCVTIHH12 pKa = 6.43VDD14 pKa = 3.58NIFWEE19 pKa = 4.34LQKK22 pKa = 10.47WNQSLTYY29 pKa = 10.54GIGQLEE35 pKa = 4.4LGLNGSTHH43 pKa = 3.99WQMYY47 pKa = 9.07FEE49 pKa = 5.24NNTAISLTQWKK60 pKa = 9.17QLLGCKK66 pKa = 9.29RR67 pKa = 11.84AHH69 pKa = 5.68VEE71 pKa = 4.13TRR73 pKa = 11.84KK74 pKa = 8.84GTALLAIEE82 pKa = 4.28YY83 pKa = 9.15CKK85 pKa = 10.71KK86 pKa = 10.65EE87 pKa = 3.99EE88 pKa = 4.19TRR90 pKa = 11.84LHH92 pKa = 6.14GPGTSFEE99 pKa = 4.27FGKK102 pKa = 7.69TTEE105 pKa = 4.11EE106 pKa = 4.04SVKK109 pKa = 10.43RR110 pKa = 11.84KK111 pKa = 10.51SPDD114 pKa = 3.14SPYY117 pKa = 11.16KK118 pKa = 10.33KK119 pKa = 10.42ALNCLSYY126 pKa = 10.95ADD128 pKa = 4.74SIAIIKK134 pKa = 10.46DD135 pKa = 3.32EE136 pKa = 4.17APRR139 pKa = 11.84DD140 pKa = 3.4YY141 pKa = 10.94VIYY144 pKa = 10.02HH145 pKa = 5.7NQVTSTLKK153 pKa = 10.9AIFKK157 pKa = 8.87PVWNKK162 pKa = 9.28TPGLVFNLEE171 pKa = 4.18PFPEE175 pKa = 4.25EE176 pKa = 3.98TLNTRR181 pKa = 11.84AVIFMGRR188 pKa = 11.84SGLGKK193 pKa = 7.31TQFAISHH200 pKa = 6.41FKK202 pKa = 10.85DD203 pKa = 3.39PLIVSHH209 pKa = 7.66IDD211 pKa = 3.39DD212 pKa = 4.6LKK214 pKa = 11.21KK215 pKa = 10.88LNPTIDD221 pKa = 4.37GIIFDD226 pKa = 5.22DD227 pKa = 4.39MNFQHH232 pKa = 7.06WPPTACIHH240 pKa = 5.92LCDD243 pKa = 3.88MEE245 pKa = 4.71LPRR248 pKa = 11.84SINVKK253 pKa = 9.84YY254 pKa = 9.03GTVEE258 pKa = 3.79IPANTPRR265 pKa = 11.84IFTSNRR271 pKa = 11.84DD272 pKa = 3.59FEE274 pKa = 5.28EE275 pKa = 4.15IWSKK279 pKa = 11.59DD280 pKa = 3.32CTFEE284 pKa = 3.75EE285 pKa = 4.43MNAIRR290 pKa = 11.84RR291 pKa = 11.84RR292 pKa = 11.84CDD294 pKa = 2.33ITTFDD299 pKa = 3.72NEE301 pKa = 4.33LYY303 pKa = 10.88
Molecular weight: 34.85 kDa
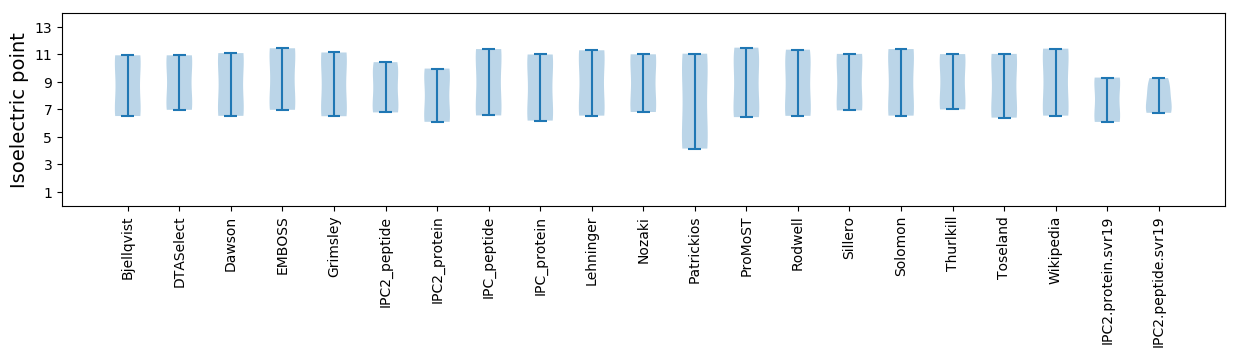
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075M3P4|A0A075M3P4_9VIRU Putative capsid protein OS=McMurdo Ice Shelf pond-associated circular DNA virus-2 OX=1521386 PE=4 SV=1
MM1 pKa = 6.79NRR3 pKa = 11.84KK4 pKa = 8.1MKK6 pKa = 9.62RR7 pKa = 11.84TGRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84TYY14 pKa = 9.05RR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 9.3RR18 pKa = 11.84NRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84SGRR25 pKa = 11.84SGRR28 pKa = 11.84TRR30 pKa = 11.84RR31 pKa = 11.84LKK33 pKa = 10.47FRR35 pKa = 11.84NTNLQSKK42 pKa = 9.63LVVFRR47 pKa = 11.84DD48 pKa = 3.41KK49 pKa = 11.11FRR51 pKa = 11.84VEE53 pKa = 4.0VQGNPNSVDD62 pKa = 3.15GVSIANSGNFNGISSAEE79 pKa = 3.94YY80 pKa = 8.81SRR82 pKa = 11.84GVRR85 pKa = 11.84EE86 pKa = 3.59CRR88 pKa = 11.84GFNRR92 pKa = 11.84IAFSGFSVSVKK103 pKa = 9.13KK104 pKa = 8.66TAEE107 pKa = 4.16CEE109 pKa = 3.93TQYY112 pKa = 10.81TVNPTVAVSAIAVNKK127 pKa = 8.38TRR129 pKa = 11.84STGRR133 pKa = 11.84LSFVTDD139 pKa = 4.18LYY141 pKa = 11.71VDD143 pKa = 4.36TTQMAANDD151 pKa = 3.86VFTEE155 pKa = 4.39TVLGVKK161 pKa = 10.25KK162 pKa = 10.75NIGFKK167 pKa = 9.99GRR169 pKa = 11.84KK170 pKa = 7.48VYY172 pKa = 9.22TYY174 pKa = 10.41KK175 pKa = 10.58VSKK178 pKa = 9.87HH179 pKa = 4.81LHH181 pKa = 5.81AGQSIKK187 pKa = 11.07VKK189 pKa = 10.67DD190 pKa = 3.97AFLTSTPPNIPWPLLLNFQSDD211 pKa = 3.86TTLTKK216 pKa = 9.39QTVPGGFFLVCDD228 pKa = 4.33TWPTAPLAPATIINTFPWHH247 pKa = 4.96TTSCQVHH254 pKa = 5.54INYY257 pKa = 9.16YY258 pKa = 8.97FKK260 pKa = 11.01VWGKK264 pKa = 8.75QATMLPLAWGG274 pKa = 3.62
MM1 pKa = 6.79NRR3 pKa = 11.84KK4 pKa = 8.1MKK6 pKa = 9.62RR7 pKa = 11.84TGRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84TYY14 pKa = 9.05RR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 9.3RR18 pKa = 11.84NRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84SGRR25 pKa = 11.84SGRR28 pKa = 11.84TRR30 pKa = 11.84RR31 pKa = 11.84LKK33 pKa = 10.47FRR35 pKa = 11.84NTNLQSKK42 pKa = 9.63LVVFRR47 pKa = 11.84DD48 pKa = 3.41KK49 pKa = 11.11FRR51 pKa = 11.84VEE53 pKa = 4.0VQGNPNSVDD62 pKa = 3.15GVSIANSGNFNGISSAEE79 pKa = 3.94YY80 pKa = 8.81SRR82 pKa = 11.84GVRR85 pKa = 11.84EE86 pKa = 3.59CRR88 pKa = 11.84GFNRR92 pKa = 11.84IAFSGFSVSVKK103 pKa = 9.13KK104 pKa = 8.66TAEE107 pKa = 4.16CEE109 pKa = 3.93TQYY112 pKa = 10.81TVNPTVAVSAIAVNKK127 pKa = 8.38TRR129 pKa = 11.84STGRR133 pKa = 11.84LSFVTDD139 pKa = 4.18LYY141 pKa = 11.71VDD143 pKa = 4.36TTQMAANDD151 pKa = 3.86VFTEE155 pKa = 4.39TVLGVKK161 pKa = 10.25KK162 pKa = 10.75NIGFKK167 pKa = 9.99GRR169 pKa = 11.84KK170 pKa = 7.48VYY172 pKa = 9.22TYY174 pKa = 10.41KK175 pKa = 10.58VSKK178 pKa = 9.87HH179 pKa = 4.81LHH181 pKa = 5.81AGQSIKK187 pKa = 11.07VKK189 pKa = 10.67DD190 pKa = 3.97AFLTSTPPNIPWPLLLNFQSDD211 pKa = 3.86TTLTKK216 pKa = 9.39QTVPGGFFLVCDD228 pKa = 4.33TWPTAPLAPATIINTFPWHH247 pKa = 4.96TTSCQVHH254 pKa = 5.54INYY257 pKa = 9.16YY258 pKa = 8.97FKK260 pKa = 11.01VWGKK264 pKa = 8.75QATMLPLAWGG274 pKa = 3.62
Molecular weight: 30.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
863 |
98 |
303 |
215.8 |
24.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.867 ± 0.375 | 2.433 ± 0.36 |
4.751 ± 0.636 | 5.446 ± 1.302 |
5.098 ± 0.378 | 6.489 ± 0.855 |
2.781 ± 0.489 | 5.91 ± 1.099 |
5.794 ± 0.861 | 6.837 ± 0.987 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.506 ± 0.203 | 5.794 ± 0.324 |
5.678 ± 1.063 | 3.824 ± 0.567 |
8.691 ± 1.843 | 5.91 ± 0.877 |
7.532 ± 1.317 | 5.91 ± 1.157 |
1.97 ± 0.158 | 2.781 ± 0.174 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
