
Microbacterium sp. CH12i
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium; unclassified Microbacterium
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
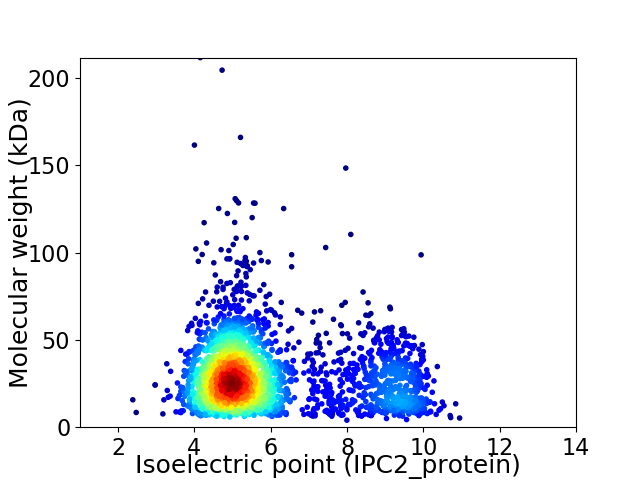
Virtual 2D-PAGE plot for 2717 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
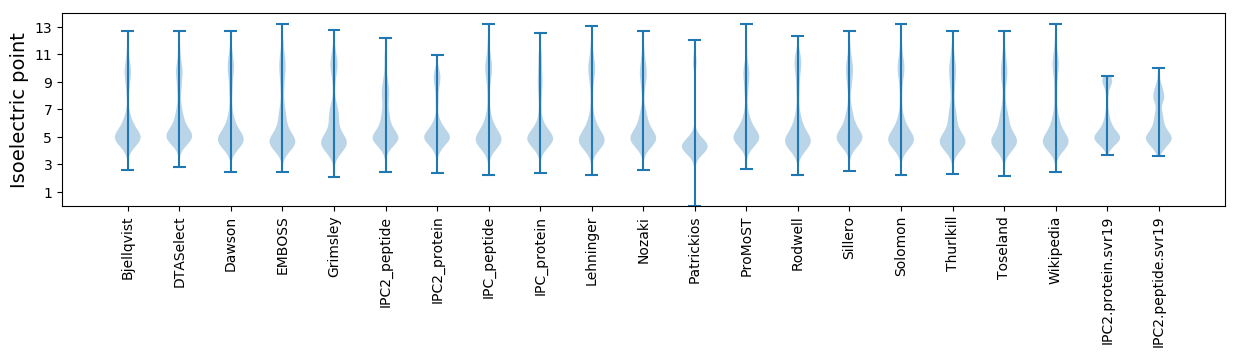
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A062VGK2|A0A062VGK2_9MICO Alanine aminopeptidase OS=Microbacterium sp. CH12i OX=1479651 GN=DC31_13595 PE=3 SV=1
MM1 pKa = 7.61NEE3 pKa = 4.03EE4 pKa = 4.1GLGDD8 pKa = 4.54EE9 pKa = 4.92IFCSLTQDD17 pKa = 4.07DD18 pKa = 4.32EE19 pKa = 5.08SGASFRR25 pKa = 11.84EE26 pKa = 4.57GFHH29 pKa = 6.61FALDD33 pKa = 3.86EE34 pKa = 4.54LDD36 pKa = 3.6LAKK39 pKa = 10.88GIDD42 pKa = 3.49VTYY45 pKa = 8.4PTGNQDD51 pKa = 3.33FVAQITQLKK60 pKa = 9.37NAGCGVVHH68 pKa = 7.0LAGTGGTLQNAAIKK82 pKa = 10.14AVQLDD87 pKa = 5.1FDD89 pKa = 4.45ATWLAPAMAVLATTATGPGAEE110 pKa = 4.36YY111 pKa = 9.66IIEE114 pKa = 4.11HH115 pKa = 5.79VRR117 pKa = 11.84VFMTGTEE124 pKa = 3.98WDD126 pKa = 3.52GDD128 pKa = 3.98QAPGQAMMQEE138 pKa = 4.55DD139 pKa = 5.15LEE141 pKa = 4.72DD142 pKa = 4.83LYY144 pKa = 10.81PDD146 pKa = 4.29YY147 pKa = 10.97IPYY150 pKa = 10.43ANSYY154 pKa = 6.17QTGYY158 pKa = 10.25LSSVTTTAILEE169 pKa = 4.07RR170 pKa = 11.84AIADD174 pKa = 3.28GDD176 pKa = 3.86LSRR179 pKa = 11.84EE180 pKa = 3.89NLMSIAASLGEE191 pKa = 4.06VDD193 pKa = 5.92DD194 pKa = 5.26LGLGGGTYY202 pKa = 10.35LYY204 pKa = 10.71GDD206 pKa = 4.61SIDD209 pKa = 3.73EE210 pKa = 4.43RR211 pKa = 11.84EE212 pKa = 4.2PTTALSVFRR221 pKa = 11.84VDD223 pKa = 3.31PAAAIGLTLEE233 pKa = 4.19KK234 pKa = 10.24FQYY237 pKa = 9.87EE238 pKa = 4.34SPLAAKK244 pKa = 10.36YY245 pKa = 9.86NAEE248 pKa = 4.0NN249 pKa = 3.48
MM1 pKa = 7.61NEE3 pKa = 4.03EE4 pKa = 4.1GLGDD8 pKa = 4.54EE9 pKa = 4.92IFCSLTQDD17 pKa = 4.07DD18 pKa = 4.32EE19 pKa = 5.08SGASFRR25 pKa = 11.84EE26 pKa = 4.57GFHH29 pKa = 6.61FALDD33 pKa = 3.86EE34 pKa = 4.54LDD36 pKa = 3.6LAKK39 pKa = 10.88GIDD42 pKa = 3.49VTYY45 pKa = 8.4PTGNQDD51 pKa = 3.33FVAQITQLKK60 pKa = 9.37NAGCGVVHH68 pKa = 7.0LAGTGGTLQNAAIKK82 pKa = 10.14AVQLDD87 pKa = 5.1FDD89 pKa = 4.45ATWLAPAMAVLATTATGPGAEE110 pKa = 4.36YY111 pKa = 9.66IIEE114 pKa = 4.11HH115 pKa = 5.79VRR117 pKa = 11.84VFMTGTEE124 pKa = 3.98WDD126 pKa = 3.52GDD128 pKa = 3.98QAPGQAMMQEE138 pKa = 4.55DD139 pKa = 5.15LEE141 pKa = 4.72DD142 pKa = 4.83LYY144 pKa = 10.81PDD146 pKa = 4.29YY147 pKa = 10.97IPYY150 pKa = 10.43ANSYY154 pKa = 6.17QTGYY158 pKa = 10.25LSSVTTTAILEE169 pKa = 4.07RR170 pKa = 11.84AIADD174 pKa = 3.28GDD176 pKa = 3.86LSRR179 pKa = 11.84EE180 pKa = 3.89NLMSIAASLGEE191 pKa = 4.06VDD193 pKa = 5.92DD194 pKa = 5.26LGLGGGTYY202 pKa = 10.35LYY204 pKa = 10.71GDD206 pKa = 4.61SIDD209 pKa = 3.73EE210 pKa = 4.43RR211 pKa = 11.84EE212 pKa = 4.2PTTALSVFRR221 pKa = 11.84VDD223 pKa = 3.31PAAAIGLTLEE233 pKa = 4.19KK234 pKa = 10.24FQYY237 pKa = 9.87EE238 pKa = 4.34SPLAAKK244 pKa = 10.36YY245 pKa = 9.86NAEE248 pKa = 4.0NN249 pKa = 3.48
Molecular weight: 26.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A062VSL9|A0A062VSL9_9MICO Exodeoxyribonuclease 7 small subunit OS=Microbacterium sp. CH12i OX=1479651 GN=xseB PE=3 SV=1
MM1 pKa = 7.51TKK3 pKa = 9.07RR4 pKa = 11.84TFQPNIRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.8KK16 pKa = 8.97HH17 pKa = 4.39GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84GILSARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.56GRR40 pKa = 11.84TEE42 pKa = 4.14LSAA45 pKa = 4.86
MM1 pKa = 7.51TKK3 pKa = 9.07RR4 pKa = 11.84TFQPNIRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.8KK16 pKa = 8.97HH17 pKa = 4.39GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84GILSARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.56GRR40 pKa = 11.84TEE42 pKa = 4.14LSAA45 pKa = 4.86
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
784182 |
37 |
2036 |
288.6 |
31.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.911 ± 0.066 | 0.534 ± 0.013 |
6.192 ± 0.042 | 5.798 ± 0.042 |
3.166 ± 0.028 | 8.539 ± 0.041 |
2.06 ± 0.023 | 5.177 ± 0.037 |
2.312 ± 0.032 | 9.982 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.988 ± 0.023 | 2.231 ± 0.025 |
5.055 ± 0.035 | 2.95 ± 0.026 |
7.019 ± 0.046 | 5.907 ± 0.033 |
6.178 ± 0.033 | 8.543 ± 0.041 |
1.496 ± 0.02 | 1.962 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
