
Capra hircus papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Phipapillomavirus; Phipapillomavirus 1
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
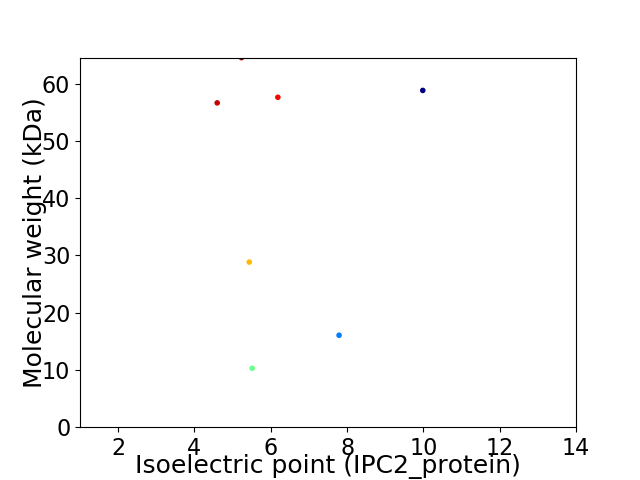
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q1I123|Q1I123_9PAPI E4 OS=Capra hircus papillomavirus 1 OX=338903 GN=E4 PE=4 SV=1
MM1 pKa = 7.01VRR3 pKa = 11.84AKK5 pKa = 9.95RR6 pKa = 11.84VKK8 pKa = 10.1RR9 pKa = 11.84ASEE12 pKa = 3.8DD13 pKa = 3.2QLYY16 pKa = 10.14RR17 pKa = 11.84DD18 pKa = 4.04CLAGRR23 pKa = 11.84EE24 pKa = 4.31CKK26 pKa = 9.91PDD28 pKa = 3.22IKK30 pKa = 10.85DD31 pKa = 3.2KK32 pKa = 11.64YY33 pKa = 9.86EE34 pKa = 4.15NNTLADD40 pKa = 5.03SILKK44 pKa = 8.89WASQFLWFGQLGIGTGRR61 pKa = 11.84GSGGSGGYY69 pKa = 7.9TRR71 pKa = 11.84LGGGGGGSRR80 pKa = 11.84TGVISRR86 pKa = 11.84PPVVIDD92 pKa = 3.76SIGPRR97 pKa = 11.84DD98 pKa = 3.79ITPIDD103 pKa = 3.83TVDD106 pKa = 3.55PTASSIVPLNEE117 pKa = 4.34GGPSVPTGEE126 pKa = 4.1IEE128 pKa = 4.75VIAEE132 pKa = 4.26TTPTTSNGALDD143 pKa = 3.87PVIVGEE149 pKa = 4.43PDD151 pKa = 3.4LGPATIDD158 pKa = 3.38VSSDD162 pKa = 3.3PTPSNPVLNTTVSSSIHH179 pKa = 5.99NNPSFDD185 pKa = 3.71AAILSSQLPGEE196 pKa = 4.29FSVPDD201 pKa = 3.43QVVITHH207 pKa = 6.47GLVGEE212 pKa = 4.63HH213 pKa = 6.44IGAFEE218 pKa = 4.59NIEE221 pKa = 4.28LDD223 pKa = 3.8TIGEE227 pKa = 4.01YY228 pKa = 10.59SGFGEE233 pKa = 4.91LEE235 pKa = 4.06VEE237 pKa = 4.44EE238 pKa = 5.3PPSSSTPQTGLSARR252 pKa = 11.84LSKK255 pKa = 10.57NLKK258 pKa = 7.75QLRR261 pKa = 11.84QRR263 pKa = 11.84LYY265 pKa = 10.6NRR267 pKa = 11.84RR268 pKa = 11.84IQQRR272 pKa = 11.84LVEE275 pKa = 4.28NQPFLTSPRR284 pKa = 11.84RR285 pKa = 11.84LVQFEE290 pKa = 4.12YY291 pKa = 10.96SNPSFQDD298 pKa = 3.3EE299 pKa = 4.35VSLEE303 pKa = 3.9FEE305 pKa = 4.01RR306 pKa = 11.84DD307 pKa = 2.96INTVVEE313 pKa = 4.28APHH316 pKa = 6.59YY317 pKa = 10.21DD318 pKa = 3.08FRR320 pKa = 11.84DD321 pKa = 3.8VVRR324 pKa = 11.84LGRR327 pKa = 11.84PEE329 pKa = 4.86SNLTDD334 pKa = 3.6SGHH337 pKa = 5.46VRR339 pKa = 11.84VSRR342 pKa = 11.84LGTLGTISTRR352 pKa = 11.84SGLIIGGQVHH362 pKa = 6.85YY363 pKa = 9.64YY364 pKa = 9.88TDD366 pKa = 4.07LSSIAADD373 pKa = 3.93TIEE376 pKa = 4.45LPSYY380 pKa = 9.52QPAMSPSTIVHH391 pKa = 6.54IDD393 pKa = 3.27SAGEE397 pKa = 4.01TFNDD401 pKa = 4.02MEE403 pKa = 4.55TSFCNVDD410 pKa = 3.16QGIEE414 pKa = 4.07EE415 pKa = 4.31VEE417 pKa = 3.92RR418 pKa = 11.84DD419 pKa = 3.84LLDD422 pKa = 3.76SASEE426 pKa = 4.09DD427 pKa = 3.75FSNSRR432 pKa = 11.84LLLSTGSDD440 pKa = 3.11IVDD443 pKa = 3.75SGLDD447 pKa = 3.42LQDD450 pKa = 2.75IFQRR454 pKa = 11.84PNLGEE459 pKa = 4.2DD460 pKa = 3.62SITVNFPTSNSIQVFNGSGAGSSIPATPIGPLGPVPIPAVLFNVLSSDD508 pKa = 4.16LLLDD512 pKa = 3.9PSLLRR517 pKa = 11.84KK518 pKa = 9.25RR519 pKa = 11.84RR520 pKa = 11.84KK521 pKa = 9.72KK522 pKa = 10.88YY523 pKa = 10.75GVFSS527 pKa = 3.89
MM1 pKa = 7.01VRR3 pKa = 11.84AKK5 pKa = 9.95RR6 pKa = 11.84VKK8 pKa = 10.1RR9 pKa = 11.84ASEE12 pKa = 3.8DD13 pKa = 3.2QLYY16 pKa = 10.14RR17 pKa = 11.84DD18 pKa = 4.04CLAGRR23 pKa = 11.84EE24 pKa = 4.31CKK26 pKa = 9.91PDD28 pKa = 3.22IKK30 pKa = 10.85DD31 pKa = 3.2KK32 pKa = 11.64YY33 pKa = 9.86EE34 pKa = 4.15NNTLADD40 pKa = 5.03SILKK44 pKa = 8.89WASQFLWFGQLGIGTGRR61 pKa = 11.84GSGGSGGYY69 pKa = 7.9TRR71 pKa = 11.84LGGGGGGSRR80 pKa = 11.84TGVISRR86 pKa = 11.84PPVVIDD92 pKa = 3.76SIGPRR97 pKa = 11.84DD98 pKa = 3.79ITPIDD103 pKa = 3.83TVDD106 pKa = 3.55PTASSIVPLNEE117 pKa = 4.34GGPSVPTGEE126 pKa = 4.1IEE128 pKa = 4.75VIAEE132 pKa = 4.26TTPTTSNGALDD143 pKa = 3.87PVIVGEE149 pKa = 4.43PDD151 pKa = 3.4LGPATIDD158 pKa = 3.38VSSDD162 pKa = 3.3PTPSNPVLNTTVSSSIHH179 pKa = 5.99NNPSFDD185 pKa = 3.71AAILSSQLPGEE196 pKa = 4.29FSVPDD201 pKa = 3.43QVVITHH207 pKa = 6.47GLVGEE212 pKa = 4.63HH213 pKa = 6.44IGAFEE218 pKa = 4.59NIEE221 pKa = 4.28LDD223 pKa = 3.8TIGEE227 pKa = 4.01YY228 pKa = 10.59SGFGEE233 pKa = 4.91LEE235 pKa = 4.06VEE237 pKa = 4.44EE238 pKa = 5.3PPSSSTPQTGLSARR252 pKa = 11.84LSKK255 pKa = 10.57NLKK258 pKa = 7.75QLRR261 pKa = 11.84QRR263 pKa = 11.84LYY265 pKa = 10.6NRR267 pKa = 11.84RR268 pKa = 11.84IQQRR272 pKa = 11.84LVEE275 pKa = 4.28NQPFLTSPRR284 pKa = 11.84RR285 pKa = 11.84LVQFEE290 pKa = 4.12YY291 pKa = 10.96SNPSFQDD298 pKa = 3.3EE299 pKa = 4.35VSLEE303 pKa = 3.9FEE305 pKa = 4.01RR306 pKa = 11.84DD307 pKa = 2.96INTVVEE313 pKa = 4.28APHH316 pKa = 6.59YY317 pKa = 10.21DD318 pKa = 3.08FRR320 pKa = 11.84DD321 pKa = 3.8VVRR324 pKa = 11.84LGRR327 pKa = 11.84PEE329 pKa = 4.86SNLTDD334 pKa = 3.6SGHH337 pKa = 5.46VRR339 pKa = 11.84VSRR342 pKa = 11.84LGTLGTISTRR352 pKa = 11.84SGLIIGGQVHH362 pKa = 6.85YY363 pKa = 9.64YY364 pKa = 9.88TDD366 pKa = 4.07LSSIAADD373 pKa = 3.93TIEE376 pKa = 4.45LPSYY380 pKa = 9.52QPAMSPSTIVHH391 pKa = 6.54IDD393 pKa = 3.27SAGEE397 pKa = 4.01TFNDD401 pKa = 4.02MEE403 pKa = 4.55TSFCNVDD410 pKa = 3.16QGIEE414 pKa = 4.07EE415 pKa = 4.31VEE417 pKa = 3.92RR418 pKa = 11.84DD419 pKa = 3.84LLDD422 pKa = 3.76SASEE426 pKa = 4.09DD427 pKa = 3.75FSNSRR432 pKa = 11.84LLLSTGSDD440 pKa = 3.11IVDD443 pKa = 3.75SGLDD447 pKa = 3.42LQDD450 pKa = 2.75IFQRR454 pKa = 11.84PNLGEE459 pKa = 4.2DD460 pKa = 3.62SITVNFPTSNSIQVFNGSGAGSSIPATPIGPLGPVPIPAVLFNVLSSDD508 pKa = 4.16LLLDD512 pKa = 3.9PSLLRR517 pKa = 11.84KK518 pKa = 9.25RR519 pKa = 11.84RR520 pKa = 11.84KK521 pKa = 9.72KK522 pKa = 10.88YY523 pKa = 10.75GVFSS527 pKa = 3.89
Molecular weight: 56.66 kDa
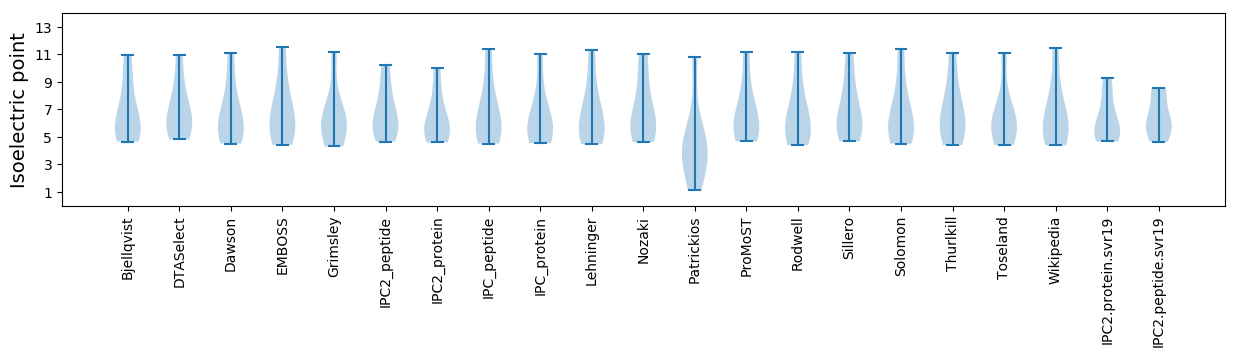
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q1I125|Q1I125_9PAPI Replication protein E1 OS=Capra hircus papillomavirus 1 OX=338903 GN=E1 PE=3 SV=1
MM1 pKa = 6.99TALEE5 pKa = 3.99ARR7 pKa = 11.84FEE9 pKa = 4.16EE10 pKa = 4.62VQEE13 pKa = 4.16KK14 pKa = 10.5LLEE17 pKa = 4.22LLEE20 pKa = 4.8RR21 pKa = 11.84NPEE24 pKa = 4.06TLDD27 pKa = 3.32EE28 pKa = 4.3QIEE31 pKa = 4.24YY32 pKa = 9.94WEE34 pKa = 4.07LVKK37 pKa = 10.44WEE39 pKa = 4.3NIYY42 pKa = 11.22YY43 pKa = 10.05NAARR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 9.57GIHH52 pKa = 6.58KK53 pKa = 10.34LGFSPVPSLQASEE66 pKa = 5.04HH67 pKa = 5.68GAKK70 pKa = 9.78EE71 pKa = 4.05AIGMKK76 pKa = 10.26LQLTNLKK83 pKa = 10.47SSGFGSEE90 pKa = 3.62KK91 pKa = 8.64WTLADD96 pKa = 3.8TSSMTYY102 pKa = 10.38LAPPKK107 pKa = 9.59HH108 pKa = 5.23TFKK111 pKa = 11.12KK112 pKa = 9.76LSKK115 pKa = 9.27PVTVTFDD122 pKa = 3.87SNSQNAVRR130 pKa = 11.84YY131 pKa = 6.2TCWSHH136 pKa = 7.28VYY138 pKa = 10.13YY139 pKa = 11.17LNDD142 pKa = 2.84NDD144 pKa = 4.79MYY146 pKa = 10.95VKK148 pKa = 10.32SYY150 pKa = 11.49SNVDD154 pKa = 3.11VHH156 pKa = 5.9GVYY159 pKa = 10.74YY160 pKa = 10.3IRR162 pKa = 11.84DD163 pKa = 3.5SRR165 pKa = 11.84KK166 pKa = 8.14QYY168 pKa = 10.47YY169 pKa = 10.79VNFEE173 pKa = 3.65KK174 pKa = 10.76DD175 pKa = 2.96ARR177 pKa = 11.84KK178 pKa = 10.03YY179 pKa = 11.04SSTGQWDD186 pKa = 3.46VSFEE190 pKa = 4.12NKK192 pKa = 9.75HH193 pKa = 5.81LSSSIASSSGHH204 pKa = 5.46TATVATPEE212 pKa = 4.25TTVTSTQASAAAASAADD229 pKa = 4.03SQEE232 pKa = 4.01EE233 pKa = 4.89TPGSTRR239 pKa = 11.84KK240 pKa = 10.01LLTATYY246 pKa = 10.43HH247 pKa = 6.77RR248 pKa = 11.84ILRR251 pKa = 11.84DD252 pKa = 3.23RR253 pKa = 11.84GSPRR257 pKa = 11.84PASRR261 pKa = 11.84SRR263 pKa = 11.84SRR265 pKa = 11.84SSTSTSSSGSSYY277 pKa = 10.66RR278 pKa = 11.84PGSRR282 pKa = 11.84RR283 pKa = 11.84SSSGRR288 pKa = 11.84GARR291 pKa = 11.84RR292 pKa = 11.84SRR294 pKa = 11.84GRR296 pKa = 11.84SWRR299 pKa = 11.84RR300 pKa = 11.84TRR302 pKa = 11.84SRR304 pKa = 11.84SQSTEE309 pKa = 3.25RR310 pKa = 11.84SRR312 pKa = 11.84GGGGGRR318 pKa = 11.84GRR320 pKa = 11.84GRR322 pKa = 11.84PRR324 pKa = 11.84RR325 pKa = 11.84SGLHH329 pKa = 5.38SRR331 pKa = 11.84PSRR334 pKa = 11.84SPAPKK339 pKa = 10.08GSGVPKK345 pKa = 10.04RR346 pKa = 11.84RR347 pKa = 11.84PSVRR351 pKa = 11.84APQSYY356 pKa = 10.24RR357 pKa = 11.84GGSQQGTPPRR367 pKa = 11.84PTAVSVASATPDD379 pKa = 3.14IPRR382 pKa = 11.84LSPWFRR388 pKa = 11.84RR389 pKa = 11.84RR390 pKa = 11.84SHH392 pKa = 5.32STPRR396 pKa = 11.84SPVRR400 pKa = 11.84APGRR404 pKa = 11.84GSPPASLGSVRR415 pKa = 11.84HH416 pKa = 5.66HH417 pKa = 6.54QSSQRR422 pKa = 11.84KK423 pKa = 8.09RR424 pKa = 11.84SRR426 pKa = 11.84SHH428 pKa = 6.09EE429 pKa = 4.07RR430 pKa = 11.84LHH432 pKa = 6.79SAVKK436 pKa = 9.95KK437 pKa = 10.04DD438 pKa = 3.4PYY440 pKa = 10.57VILVRR445 pKa = 11.84GKK447 pKa = 9.58PNQLKK452 pKa = 8.33CWRR455 pKa = 11.84WRR457 pKa = 11.84LHH459 pKa = 4.34NRR461 pKa = 11.84ARR463 pKa = 11.84RR464 pKa = 11.84VPFRR468 pKa = 11.84CFSTTFSWVSLVQGEE483 pKa = 4.15KK484 pKa = 10.53SIANRR489 pKa = 11.84MLVVFDD495 pKa = 3.63TRR497 pKa = 11.84QDD499 pKa = 3.33RR500 pKa = 11.84EE501 pKa = 4.36LFLHH505 pKa = 6.8RR506 pKa = 11.84VTFPPGVTWCYY517 pKa = 11.14GSLDD521 pKa = 4.28CII523 pKa = 4.39
MM1 pKa = 6.99TALEE5 pKa = 3.99ARR7 pKa = 11.84FEE9 pKa = 4.16EE10 pKa = 4.62VQEE13 pKa = 4.16KK14 pKa = 10.5LLEE17 pKa = 4.22LLEE20 pKa = 4.8RR21 pKa = 11.84NPEE24 pKa = 4.06TLDD27 pKa = 3.32EE28 pKa = 4.3QIEE31 pKa = 4.24YY32 pKa = 9.94WEE34 pKa = 4.07LVKK37 pKa = 10.44WEE39 pKa = 4.3NIYY42 pKa = 11.22YY43 pKa = 10.05NAARR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 9.57GIHH52 pKa = 6.58KK53 pKa = 10.34LGFSPVPSLQASEE66 pKa = 5.04HH67 pKa = 5.68GAKK70 pKa = 9.78EE71 pKa = 4.05AIGMKK76 pKa = 10.26LQLTNLKK83 pKa = 10.47SSGFGSEE90 pKa = 3.62KK91 pKa = 8.64WTLADD96 pKa = 3.8TSSMTYY102 pKa = 10.38LAPPKK107 pKa = 9.59HH108 pKa = 5.23TFKK111 pKa = 11.12KK112 pKa = 9.76LSKK115 pKa = 9.27PVTVTFDD122 pKa = 3.87SNSQNAVRR130 pKa = 11.84YY131 pKa = 6.2TCWSHH136 pKa = 7.28VYY138 pKa = 10.13YY139 pKa = 11.17LNDD142 pKa = 2.84NDD144 pKa = 4.79MYY146 pKa = 10.95VKK148 pKa = 10.32SYY150 pKa = 11.49SNVDD154 pKa = 3.11VHH156 pKa = 5.9GVYY159 pKa = 10.74YY160 pKa = 10.3IRR162 pKa = 11.84DD163 pKa = 3.5SRR165 pKa = 11.84KK166 pKa = 8.14QYY168 pKa = 10.47YY169 pKa = 10.79VNFEE173 pKa = 3.65KK174 pKa = 10.76DD175 pKa = 2.96ARR177 pKa = 11.84KK178 pKa = 10.03YY179 pKa = 11.04SSTGQWDD186 pKa = 3.46VSFEE190 pKa = 4.12NKK192 pKa = 9.75HH193 pKa = 5.81LSSSIASSSGHH204 pKa = 5.46TATVATPEE212 pKa = 4.25TTVTSTQASAAAASAADD229 pKa = 4.03SQEE232 pKa = 4.01EE233 pKa = 4.89TPGSTRR239 pKa = 11.84KK240 pKa = 10.01LLTATYY246 pKa = 10.43HH247 pKa = 6.77RR248 pKa = 11.84ILRR251 pKa = 11.84DD252 pKa = 3.23RR253 pKa = 11.84GSPRR257 pKa = 11.84PASRR261 pKa = 11.84SRR263 pKa = 11.84SRR265 pKa = 11.84SSTSTSSSGSSYY277 pKa = 10.66RR278 pKa = 11.84PGSRR282 pKa = 11.84RR283 pKa = 11.84SSSGRR288 pKa = 11.84GARR291 pKa = 11.84RR292 pKa = 11.84SRR294 pKa = 11.84GRR296 pKa = 11.84SWRR299 pKa = 11.84RR300 pKa = 11.84TRR302 pKa = 11.84SRR304 pKa = 11.84SQSTEE309 pKa = 3.25RR310 pKa = 11.84SRR312 pKa = 11.84GGGGGRR318 pKa = 11.84GRR320 pKa = 11.84GRR322 pKa = 11.84PRR324 pKa = 11.84RR325 pKa = 11.84SGLHH329 pKa = 5.38SRR331 pKa = 11.84PSRR334 pKa = 11.84SPAPKK339 pKa = 10.08GSGVPKK345 pKa = 10.04RR346 pKa = 11.84RR347 pKa = 11.84PSVRR351 pKa = 11.84APQSYY356 pKa = 10.24RR357 pKa = 11.84GGSQQGTPPRR367 pKa = 11.84PTAVSVASATPDD379 pKa = 3.14IPRR382 pKa = 11.84LSPWFRR388 pKa = 11.84RR389 pKa = 11.84RR390 pKa = 11.84SHH392 pKa = 5.32STPRR396 pKa = 11.84SPVRR400 pKa = 11.84APGRR404 pKa = 11.84GSPPASLGSVRR415 pKa = 11.84HH416 pKa = 5.66HH417 pKa = 6.54QSSQRR422 pKa = 11.84KK423 pKa = 8.09RR424 pKa = 11.84SRR426 pKa = 11.84SHH428 pKa = 6.09EE429 pKa = 4.07RR430 pKa = 11.84LHH432 pKa = 6.79SAVKK436 pKa = 9.95KK437 pKa = 10.04DD438 pKa = 3.4PYY440 pKa = 10.57VILVRR445 pKa = 11.84GKK447 pKa = 9.58PNQLKK452 pKa = 8.33CWRR455 pKa = 11.84WRR457 pKa = 11.84LHH459 pKa = 4.34NRR461 pKa = 11.84ARR463 pKa = 11.84RR464 pKa = 11.84VPFRR468 pKa = 11.84CFSTTFSWVSLVQGEE483 pKa = 4.15KK484 pKa = 10.53SIANRR489 pKa = 11.84MLVVFDD495 pKa = 3.63TRR497 pKa = 11.84QDD499 pKa = 3.33RR500 pKa = 11.84EE501 pKa = 4.36LFLHH505 pKa = 6.8RR506 pKa = 11.84VTFPPGVTWCYY517 pKa = 11.14GSLDD521 pKa = 4.28CII523 pKa = 4.39
Molecular weight: 58.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2611 |
92 |
566 |
373.0 |
41.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.4 ± 0.662 | 2.106 ± 0.6 |
6.09 ± 0.912 | 5.668 ± 0.349 |
4.098 ± 0.562 | 6.817 ± 0.917 |
2.375 ± 0.327 | 4.251 ± 0.76 |
4.787 ± 0.793 | 8.771 ± 0.727 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.379 ± 0.252 | 3.983 ± 0.77 |
7.085 ± 1.56 | 4.787 ± 0.658 |
7.124 ± 1.416 | 9.307 ± 1.489 |
5.553 ± 0.514 | 5.898 ± 0.57 |
1.34 ± 0.3 | 3.179 ± 0.414 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
