
Chagres virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Chagres phlebovirus
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
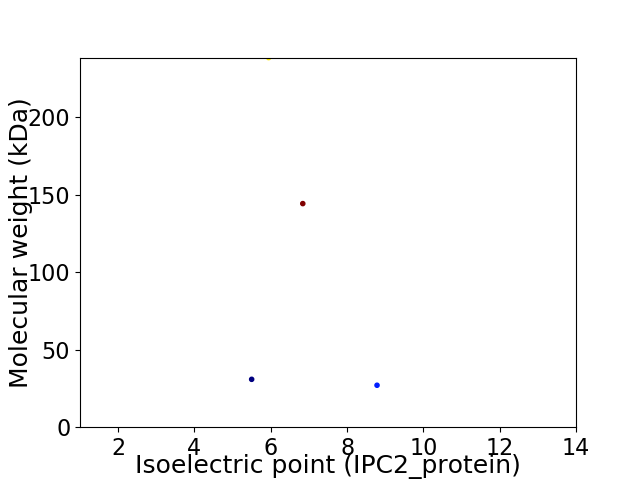
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I1T327|I1T327_9VIRU Nonstructural protein OS=Chagres virus OX=629727 PE=4 SV=1
MM1 pKa = 7.65NYY3 pKa = 10.03YY4 pKa = 9.43ATDD7 pKa = 3.36IPLVSFSACQLRR19 pKa = 11.84RR20 pKa = 11.84IQVDD24 pKa = 3.46YY25 pKa = 11.35IPFNKK30 pKa = 9.62QATEE34 pKa = 3.64PHH36 pKa = 6.05SNYY39 pKa = 10.58SGMEE43 pKa = 4.06FPVTSYY49 pKa = 9.65TQVPGIRR56 pKa = 11.84SKK58 pKa = 11.28LQDD61 pKa = 4.15FYY63 pKa = 12.05NHH65 pKa = 5.96NQLPVSWGSGRR76 pKa = 11.84PGVVNDD82 pKa = 3.97SSSKK86 pKa = 9.8YY87 pKa = 9.42EE88 pKa = 3.92SLIKK92 pKa = 10.29EE93 pKa = 3.81ISKK96 pKa = 11.05LEE98 pKa = 3.78LRR100 pKa = 11.84DD101 pKa = 3.75VVKK104 pKa = 10.89HH105 pKa = 5.37NEE107 pKa = 3.8PNIRR111 pKa = 11.84RR112 pKa = 11.84ALCWPYY118 pKa = 11.04SYY120 pKa = 8.1PTLIFFDD127 pKa = 3.62LCGNKK132 pKa = 10.21DD133 pKa = 3.71SMGPWMFKK141 pKa = 9.82GAAMTSFMRR150 pKa = 11.84AGKK153 pKa = 9.48CAQIDD158 pKa = 3.55RR159 pKa = 11.84SLVNFHH165 pKa = 6.6RR166 pKa = 11.84MIVRR170 pKa = 11.84EE171 pKa = 4.3AIEE174 pKa = 4.2LGEE177 pKa = 4.27NEE179 pKa = 5.01AEE181 pKa = 4.2FTGKK185 pKa = 10.16DD186 pKa = 3.43IIMEE190 pKa = 4.5CCSVICRR197 pKa = 11.84RR198 pKa = 11.84LLRR201 pKa = 11.84AYY203 pKa = 9.69PFDD206 pKa = 3.49RR207 pKa = 11.84TYY209 pKa = 11.47DD210 pKa = 3.67KK211 pKa = 10.9PHH213 pKa = 6.34SRR215 pKa = 11.84LLTILEE221 pKa = 4.57DD222 pKa = 3.95FDD224 pKa = 4.07NHH226 pKa = 6.64FKK228 pKa = 10.78FDD230 pKa = 3.83YY231 pKa = 10.96GEE233 pKa = 4.14SSEE236 pKa = 4.78LRR238 pKa = 11.84VAVDD242 pKa = 3.39EE243 pKa = 4.8FDD245 pKa = 3.06IHH247 pKa = 5.08MKK249 pKa = 10.22RR250 pKa = 11.84RR251 pKa = 11.84EE252 pKa = 4.07AEE254 pKa = 3.92EE255 pKa = 4.36TFWGSDD261 pKa = 3.49FTDD264 pKa = 3.28SDD266 pKa = 3.72
MM1 pKa = 7.65NYY3 pKa = 10.03YY4 pKa = 9.43ATDD7 pKa = 3.36IPLVSFSACQLRR19 pKa = 11.84RR20 pKa = 11.84IQVDD24 pKa = 3.46YY25 pKa = 11.35IPFNKK30 pKa = 9.62QATEE34 pKa = 3.64PHH36 pKa = 6.05SNYY39 pKa = 10.58SGMEE43 pKa = 4.06FPVTSYY49 pKa = 9.65TQVPGIRR56 pKa = 11.84SKK58 pKa = 11.28LQDD61 pKa = 4.15FYY63 pKa = 12.05NHH65 pKa = 5.96NQLPVSWGSGRR76 pKa = 11.84PGVVNDD82 pKa = 3.97SSSKK86 pKa = 9.8YY87 pKa = 9.42EE88 pKa = 3.92SLIKK92 pKa = 10.29EE93 pKa = 3.81ISKK96 pKa = 11.05LEE98 pKa = 3.78LRR100 pKa = 11.84DD101 pKa = 3.75VVKK104 pKa = 10.89HH105 pKa = 5.37NEE107 pKa = 3.8PNIRR111 pKa = 11.84RR112 pKa = 11.84ALCWPYY118 pKa = 11.04SYY120 pKa = 8.1PTLIFFDD127 pKa = 3.62LCGNKK132 pKa = 10.21DD133 pKa = 3.71SMGPWMFKK141 pKa = 9.82GAAMTSFMRR150 pKa = 11.84AGKK153 pKa = 9.48CAQIDD158 pKa = 3.55RR159 pKa = 11.84SLVNFHH165 pKa = 6.6RR166 pKa = 11.84MIVRR170 pKa = 11.84EE171 pKa = 4.3AIEE174 pKa = 4.2LGEE177 pKa = 4.27NEE179 pKa = 5.01AEE181 pKa = 4.2FTGKK185 pKa = 10.16DD186 pKa = 3.43IIMEE190 pKa = 4.5CCSVICRR197 pKa = 11.84RR198 pKa = 11.84LLRR201 pKa = 11.84AYY203 pKa = 9.69PFDD206 pKa = 3.49RR207 pKa = 11.84TYY209 pKa = 11.47DD210 pKa = 3.67KK211 pKa = 10.9PHH213 pKa = 6.34SRR215 pKa = 11.84LLTILEE221 pKa = 4.57DD222 pKa = 3.95FDD224 pKa = 4.07NHH226 pKa = 6.64FKK228 pKa = 10.78FDD230 pKa = 3.83YY231 pKa = 10.96GEE233 pKa = 4.14SSEE236 pKa = 4.78LRR238 pKa = 11.84VAVDD242 pKa = 3.39EE243 pKa = 4.8FDD245 pKa = 3.06IHH247 pKa = 5.08MKK249 pKa = 10.22RR250 pKa = 11.84RR251 pKa = 11.84EE252 pKa = 4.07AEE254 pKa = 3.92EE255 pKa = 4.36TFWGSDD261 pKa = 3.49FTDD264 pKa = 3.28SDD266 pKa = 3.72
Molecular weight: 30.9 kDa
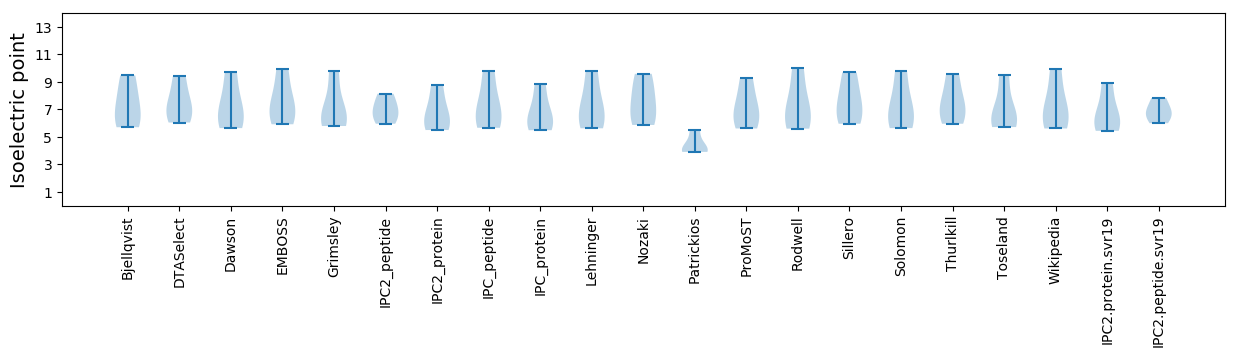
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I1T327|I1T327_9VIRU Nonstructural protein OS=Chagres virus OX=629727 PE=4 SV=1
MM1 pKa = 7.86ANYY4 pKa = 10.15QDD6 pKa = 3.55IAVEE10 pKa = 4.11FAGDD14 pKa = 3.99LPTEE18 pKa = 4.62AIIRR22 pKa = 11.84TWVEE26 pKa = 3.31EE27 pKa = 3.76FAYY30 pKa = 10.44QGFDD34 pKa = 2.76ASQVLRR40 pKa = 11.84ILAEE44 pKa = 3.86RR45 pKa = 11.84GGANWRR51 pKa = 11.84EE52 pKa = 4.2DD53 pKa = 3.62AKK55 pKa = 11.53KK56 pKa = 10.09MIILSLTRR64 pKa = 11.84GNKK67 pKa = 7.45PSKK70 pKa = 9.53MMEE73 pKa = 4.03RR74 pKa = 11.84MSEE77 pKa = 3.8AGKK80 pKa = 9.13ATVSSLMTRR89 pKa = 11.84YY90 pKa = 9.42NLKK93 pKa = 10.32SGNPGRR99 pKa = 11.84NDD101 pKa = 3.09LTLSRR106 pKa = 11.84VATALAGWTCQAIPIVQDD124 pKa = 3.51FLPVTGVVMDD134 pKa = 4.81GLSPAYY140 pKa = 8.45PRR142 pKa = 11.84PMMHH146 pKa = 7.03PCFAGLIDD154 pKa = 4.25PSLPKK159 pKa = 9.68KK160 pKa = 9.13TVDD163 pKa = 3.35EE164 pKa = 4.24LVAAFSLYY172 pKa = 9.55MLQFSKK178 pKa = 9.92TINPSLRR185 pKa = 11.84AAKK188 pKa = 10.25RR189 pKa = 11.84EE190 pKa = 4.15DD191 pKa = 3.68IISSFRR197 pKa = 11.84QPMQAAINSTFLTGAQRR214 pKa = 11.84RR215 pKa = 11.84QFLRR219 pKa = 11.84TLGVVNDD226 pKa = 4.02NLEE229 pKa = 4.09PSQNVVAAARR239 pKa = 11.84VFRR242 pKa = 11.84GTGVV246 pKa = 2.84
MM1 pKa = 7.86ANYY4 pKa = 10.15QDD6 pKa = 3.55IAVEE10 pKa = 4.11FAGDD14 pKa = 3.99LPTEE18 pKa = 4.62AIIRR22 pKa = 11.84TWVEE26 pKa = 3.31EE27 pKa = 3.76FAYY30 pKa = 10.44QGFDD34 pKa = 2.76ASQVLRR40 pKa = 11.84ILAEE44 pKa = 3.86RR45 pKa = 11.84GGANWRR51 pKa = 11.84EE52 pKa = 4.2DD53 pKa = 3.62AKK55 pKa = 11.53KK56 pKa = 10.09MIILSLTRR64 pKa = 11.84GNKK67 pKa = 7.45PSKK70 pKa = 9.53MMEE73 pKa = 4.03RR74 pKa = 11.84MSEE77 pKa = 3.8AGKK80 pKa = 9.13ATVSSLMTRR89 pKa = 11.84YY90 pKa = 9.42NLKK93 pKa = 10.32SGNPGRR99 pKa = 11.84NDD101 pKa = 3.09LTLSRR106 pKa = 11.84VATALAGWTCQAIPIVQDD124 pKa = 3.51FLPVTGVVMDD134 pKa = 4.81GLSPAYY140 pKa = 8.45PRR142 pKa = 11.84PMMHH146 pKa = 7.03PCFAGLIDD154 pKa = 4.25PSLPKK159 pKa = 9.68KK160 pKa = 9.13TVDD163 pKa = 3.35EE164 pKa = 4.24LVAAFSLYY172 pKa = 9.55MLQFSKK178 pKa = 9.92TINPSLRR185 pKa = 11.84AAKK188 pKa = 10.25RR189 pKa = 11.84EE190 pKa = 4.15DD191 pKa = 3.68IISSFRR197 pKa = 11.84QPMQAAINSTFLTGAQRR214 pKa = 11.84RR215 pKa = 11.84QFLRR219 pKa = 11.84TLGVVNDD226 pKa = 4.02NLEE229 pKa = 4.09PSQNVVAAARR239 pKa = 11.84VFRR242 pKa = 11.84GTGVV246 pKa = 2.84
Molecular weight: 27.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3909 |
246 |
2093 |
977.3 |
110.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.347 ± 0.696 | 2.814 ± 0.663 |
5.475 ± 0.43 | 6.395 ± 0.527 |
4.528 ± 0.434 | 5.602 ± 0.186 |
2.584 ± 0.287 | 7.061 ± 0.377 |
6.344 ± 0.462 | 8.647 ± 0.274 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.223 ± 0.165 | 4.63 ± 0.04 |
3.837 ± 0.332 | 3.377 ± 0.23 |
5.091 ± 0.554 | 9.849 ± 0.787 |
5.04 ± 0.169 | 6.012 ± 0.295 |
1.228 ± 0.1 | 2.916 ± 0.256 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
