
Human feces smacovirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; Porprismacovirus humas1
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
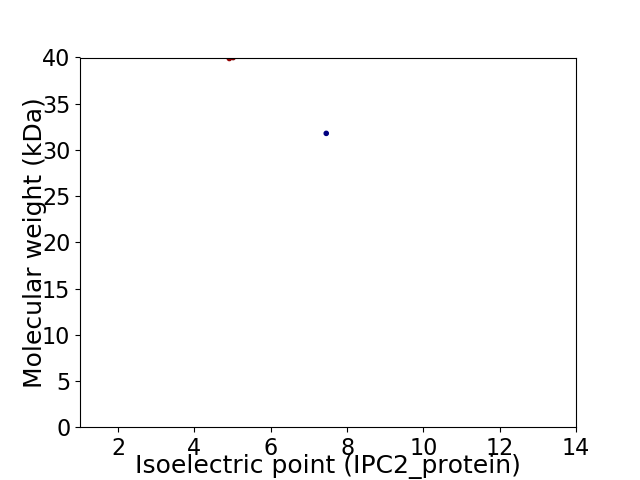
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
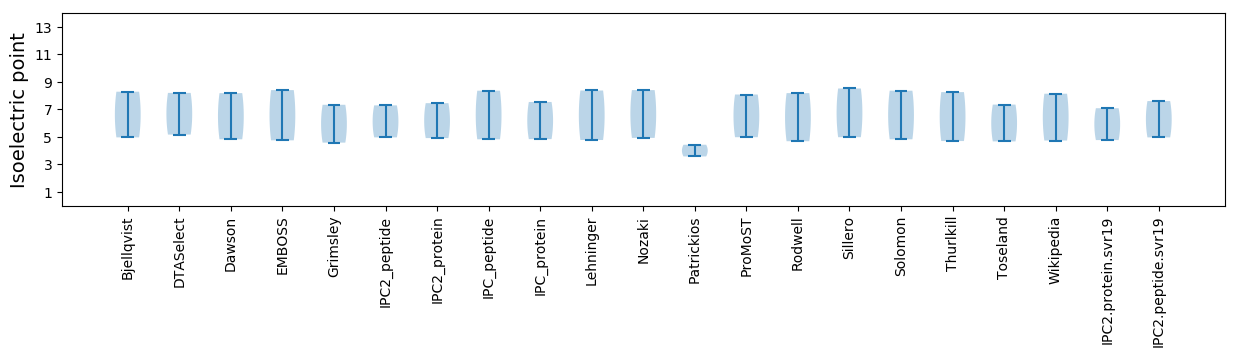
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A142J7I2|A0A142J7I2_9VIRU Replication associated protein OS=Human feces smacovirus 2 OX=1820158 PE=4 SV=1
MM1 pKa = 7.01FVKK4 pKa = 10.65VSEE7 pKa = 4.5TYY9 pKa = 10.75DD10 pKa = 3.33LSTKK14 pKa = 8.7VGKK17 pKa = 9.38MGMLGIHH24 pKa = 6.04TPDD27 pKa = 2.9GRR29 pKa = 11.84LVYY32 pKa = 10.92NMWKK36 pKa = 10.35GFYY39 pKa = 9.89QNYY42 pKa = 8.72RR43 pKa = 11.84KK44 pKa = 8.85MRR46 pKa = 11.84FVSCDD51 pKa = 3.03VALACASMLPADD63 pKa = 4.27PLQVGVEE70 pKa = 4.11AGDD73 pKa = 3.85IAPQDD78 pKa = 3.63MFNPILYY85 pKa = 9.77KK86 pKa = 10.6AVSNDD91 pKa = 3.59SMTLLLNRR99 pKa = 11.84IYY101 pKa = 10.98AGAHH105 pKa = 6.18KK106 pKa = 9.93EE107 pKa = 4.25SNPDD111 pKa = 3.04WVGKK115 pKa = 10.55NSVTDD120 pKa = 3.99DD121 pKa = 3.72NEE123 pKa = 4.1TSFVYY128 pKa = 10.64DD129 pKa = 2.99STRR132 pKa = 11.84EE133 pKa = 3.68VDD135 pKa = 3.53QFAMYY140 pKa = 10.44YY141 pKa = 10.68GLLSDD146 pKa = 3.69TSGWKK151 pKa = 9.93KK152 pKa = 10.98AMPQAGLSMSRR163 pKa = 11.84LRR165 pKa = 11.84PITWSLLASQGQPGVCGGFSNSVTYY190 pKa = 10.17PSDD193 pKa = 3.42GSSAGSSGNVVTSKK207 pKa = 9.33TRR209 pKa = 11.84EE210 pKa = 3.67LGRR213 pKa = 11.84FLRR216 pKa = 11.84GPACVMPAIDD226 pKa = 4.05TMVMTSDD233 pKa = 4.45GSCVNSSSEE242 pKa = 3.79QTNLVVTQAEE252 pKa = 4.33LPYY255 pKa = 10.94VNTSDD260 pKa = 3.32IQLGPGVASTVVLPNLDD277 pKa = 3.63VPDD280 pKa = 4.63CFVAAIILPPAKK292 pKa = 10.32LSRR295 pKa = 11.84LYY297 pKa = 10.97YY298 pKa = 9.4RR299 pKa = 11.84LKK301 pKa = 9.65VTWTVEE307 pKa = 4.01FFGPRR312 pKa = 11.84PLTDD316 pKa = 3.12LTNWYY321 pKa = 10.05GLALQGTQSYY331 pKa = 10.52GSDD334 pKa = 3.68YY335 pKa = 11.07VDD337 pKa = 3.4QAALITSGSTAGKK350 pKa = 9.6QAMVDD355 pKa = 3.44AGDD358 pKa = 4.21SEE360 pKa = 4.39IQKK363 pKa = 10.04VMEE366 pKa = 4.46SASS369 pKa = 3.36
MM1 pKa = 7.01FVKK4 pKa = 10.65VSEE7 pKa = 4.5TYY9 pKa = 10.75DD10 pKa = 3.33LSTKK14 pKa = 8.7VGKK17 pKa = 9.38MGMLGIHH24 pKa = 6.04TPDD27 pKa = 2.9GRR29 pKa = 11.84LVYY32 pKa = 10.92NMWKK36 pKa = 10.35GFYY39 pKa = 9.89QNYY42 pKa = 8.72RR43 pKa = 11.84KK44 pKa = 8.85MRR46 pKa = 11.84FVSCDD51 pKa = 3.03VALACASMLPADD63 pKa = 4.27PLQVGVEE70 pKa = 4.11AGDD73 pKa = 3.85IAPQDD78 pKa = 3.63MFNPILYY85 pKa = 9.77KK86 pKa = 10.6AVSNDD91 pKa = 3.59SMTLLLNRR99 pKa = 11.84IYY101 pKa = 10.98AGAHH105 pKa = 6.18KK106 pKa = 9.93EE107 pKa = 4.25SNPDD111 pKa = 3.04WVGKK115 pKa = 10.55NSVTDD120 pKa = 3.99DD121 pKa = 3.72NEE123 pKa = 4.1TSFVYY128 pKa = 10.64DD129 pKa = 2.99STRR132 pKa = 11.84EE133 pKa = 3.68VDD135 pKa = 3.53QFAMYY140 pKa = 10.44YY141 pKa = 10.68GLLSDD146 pKa = 3.69TSGWKK151 pKa = 9.93KK152 pKa = 10.98AMPQAGLSMSRR163 pKa = 11.84LRR165 pKa = 11.84PITWSLLASQGQPGVCGGFSNSVTYY190 pKa = 10.17PSDD193 pKa = 3.42GSSAGSSGNVVTSKK207 pKa = 9.33TRR209 pKa = 11.84EE210 pKa = 3.67LGRR213 pKa = 11.84FLRR216 pKa = 11.84GPACVMPAIDD226 pKa = 4.05TMVMTSDD233 pKa = 4.45GSCVNSSSEE242 pKa = 3.79QTNLVVTQAEE252 pKa = 4.33LPYY255 pKa = 10.94VNTSDD260 pKa = 3.32IQLGPGVASTVVLPNLDD277 pKa = 3.63VPDD280 pKa = 4.63CFVAAIILPPAKK292 pKa = 10.32LSRR295 pKa = 11.84LYY297 pKa = 10.97YY298 pKa = 9.4RR299 pKa = 11.84LKK301 pKa = 9.65VTWTVEE307 pKa = 4.01FFGPRR312 pKa = 11.84PLTDD316 pKa = 3.12LTNWYY321 pKa = 10.05GLALQGTQSYY331 pKa = 10.52GSDD334 pKa = 3.68YY335 pKa = 11.07VDD337 pKa = 3.4QAALITSGSTAGKK350 pKa = 9.6QAMVDD355 pKa = 3.44AGDD358 pKa = 4.21SEE360 pKa = 4.39IQKK363 pKa = 10.04VMEE366 pKa = 4.46SASS369 pKa = 3.36
Molecular weight: 39.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A142J7I2|A0A142J7I2_9VIRU Replication associated protein OS=Human feces smacovirus 2 OX=1820158 PE=4 SV=1
MM1 pKa = 8.19DD2 pKa = 3.6IMATIGRR9 pKa = 11.84TGYY12 pKa = 10.62SGFAEE17 pKa = 3.65IMAIIRR23 pKa = 11.84EE24 pKa = 4.09LDD26 pKa = 2.9IHH28 pKa = 6.26KK29 pKa = 9.73WIIGAEE35 pKa = 4.12VGAGGYY41 pKa = 7.23QHH43 pKa = 6.13WQCRR47 pKa = 11.84FKK49 pKa = 10.83TRR51 pKa = 11.84LSPEE55 pKa = 3.61EE56 pKa = 4.26RR57 pKa = 11.84IEE59 pKa = 4.13GLEE62 pKa = 3.97WDD64 pKa = 4.68LKK66 pKa = 9.38TATVKK71 pKa = 10.01QKK73 pKa = 10.67KK74 pKa = 7.91RR75 pKa = 11.84TVGPAEE81 pKa = 3.89QMTEE85 pKa = 3.74KK86 pKa = 10.53LRR88 pKa = 11.84ARR90 pKa = 11.84IGDD93 pKa = 3.65GPSIYY98 pKa = 9.87TAEE101 pKa = 4.86CSDD104 pKa = 2.93NWEE107 pKa = 4.19YY108 pKa = 10.45EE109 pKa = 4.26AKK111 pKa = 10.34EE112 pKa = 4.05GAYY115 pKa = 9.87LASWDD120 pKa = 3.8TMKK123 pKa = 10.36VRR125 pKa = 11.84STRR128 pKa = 11.84FGKK131 pKa = 9.73PSWQQEE137 pKa = 4.04VALQALRR144 pKa = 11.84DD145 pKa = 3.87TNDD148 pKa = 3.24RR149 pKa = 11.84QVVVWYY155 pKa = 9.73DD156 pKa = 3.32PQGNIGKK163 pKa = 8.92SWLTNHH169 pKa = 7.47LYY171 pKa = 10.28EE172 pKa = 5.34QGLAYY177 pKa = 9.93CIPATMNNIEE187 pKa = 4.64GIIKK191 pKa = 8.85TVASLAVKK199 pKa = 10.12DD200 pKa = 4.25RR201 pKa = 11.84DD202 pKa = 3.15MGYY205 pKa = 9.56PPRR208 pKa = 11.84PYY210 pKa = 10.55VIIDD214 pKa = 3.66IPRR217 pKa = 11.84SWKK220 pKa = 9.68WSEE223 pKa = 3.9QLYY226 pKa = 8.31TAIEE230 pKa = 4.32SIKK233 pKa = 10.66DD234 pKa = 3.62GLIMDD239 pKa = 5.0PRR241 pKa = 11.84YY242 pKa = 9.77SAQPINIRR250 pKa = 11.84GVKK253 pKa = 9.21VMVMTNTMPKK263 pKa = 8.96IDD265 pKa = 4.66KK266 pKa = 10.18LSSDD270 pKa = 3.03RR271 pKa = 11.84WVIHH275 pKa = 5.71TNPP278 pKa = 3.61
MM1 pKa = 8.19DD2 pKa = 3.6IMATIGRR9 pKa = 11.84TGYY12 pKa = 10.62SGFAEE17 pKa = 3.65IMAIIRR23 pKa = 11.84EE24 pKa = 4.09LDD26 pKa = 2.9IHH28 pKa = 6.26KK29 pKa = 9.73WIIGAEE35 pKa = 4.12VGAGGYY41 pKa = 7.23QHH43 pKa = 6.13WQCRR47 pKa = 11.84FKK49 pKa = 10.83TRR51 pKa = 11.84LSPEE55 pKa = 3.61EE56 pKa = 4.26RR57 pKa = 11.84IEE59 pKa = 4.13GLEE62 pKa = 3.97WDD64 pKa = 4.68LKK66 pKa = 9.38TATVKK71 pKa = 10.01QKK73 pKa = 10.67KK74 pKa = 7.91RR75 pKa = 11.84TVGPAEE81 pKa = 3.89QMTEE85 pKa = 3.74KK86 pKa = 10.53LRR88 pKa = 11.84ARR90 pKa = 11.84IGDD93 pKa = 3.65GPSIYY98 pKa = 9.87TAEE101 pKa = 4.86CSDD104 pKa = 2.93NWEE107 pKa = 4.19YY108 pKa = 10.45EE109 pKa = 4.26AKK111 pKa = 10.34EE112 pKa = 4.05GAYY115 pKa = 9.87LASWDD120 pKa = 3.8TMKK123 pKa = 10.36VRR125 pKa = 11.84STRR128 pKa = 11.84FGKK131 pKa = 9.73PSWQQEE137 pKa = 4.04VALQALRR144 pKa = 11.84DD145 pKa = 3.87TNDD148 pKa = 3.24RR149 pKa = 11.84QVVVWYY155 pKa = 9.73DD156 pKa = 3.32PQGNIGKK163 pKa = 8.92SWLTNHH169 pKa = 7.47LYY171 pKa = 10.28EE172 pKa = 5.34QGLAYY177 pKa = 9.93CIPATMNNIEE187 pKa = 4.64GIIKK191 pKa = 8.85TVASLAVKK199 pKa = 10.12DD200 pKa = 4.25RR201 pKa = 11.84DD202 pKa = 3.15MGYY205 pKa = 9.56PPRR208 pKa = 11.84PYY210 pKa = 10.55VIIDD214 pKa = 3.66IPRR217 pKa = 11.84SWKK220 pKa = 9.68WSEE223 pKa = 3.9QLYY226 pKa = 8.31TAIEE230 pKa = 4.32SIKK233 pKa = 10.66DD234 pKa = 3.62GLIMDD239 pKa = 5.0PRR241 pKa = 11.84YY242 pKa = 9.77SAQPINIRR250 pKa = 11.84GVKK253 pKa = 9.21VMVMTNTMPKK263 pKa = 8.96IDD265 pKa = 4.66KK266 pKa = 10.18LSSDD270 pKa = 3.03RR271 pKa = 11.84WVIHH275 pKa = 5.71TNPP278 pKa = 3.61
Molecular weight: 31.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
647 |
278 |
369 |
323.5 |
35.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.419 ± 0.12 | 1.391 ± 0.167 |
6.182 ± 0.228 | 4.482 ± 1.064 |
2.164 ± 0.579 | 7.883 ± 0.368 |
0.927 ± 0.273 | 5.719 ± 1.941 |
5.1 ± 0.734 | 7.264 ± 0.998 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.173 ± 0.116 | 3.709 ± 0.252 |
5.255 ± 0.117 | 4.173 ± 0.077 |
4.791 ± 0.899 | 8.346 ± 1.576 |
6.646 ± 0.091 | 7.419 ± 1.081 |
2.628 ± 0.71 | 4.328 ± 0.006 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
