
Feline calicivirus (strain Cat/United States/Urbana/1960) (FCV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Picornavirales; Caliciviridae; Vesivirus; Feline calicivirus
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
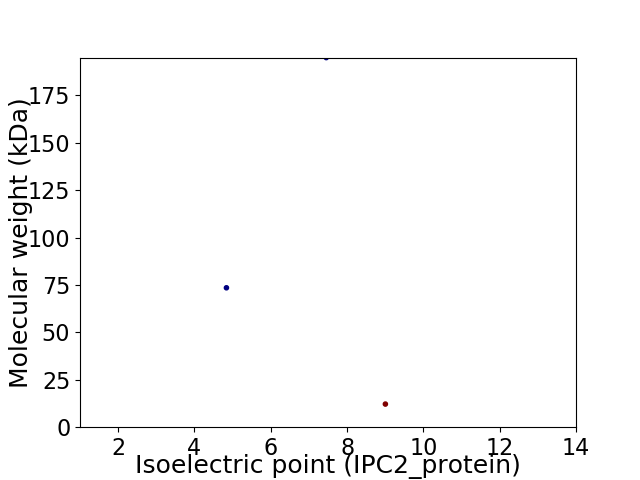
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q66916|VP2_FCVUR Protein VP2 OS=Feline calicivirus (strain Cat/United States/Urbana/1960) OX=292349 GN=ORF3 PE=1 SV=1
MM1 pKa = 7.93CSTCANVLKK10 pKa = 10.53YY11 pKa = 10.45YY12 pKa = 10.68NWDD15 pKa = 3.24PHH17 pKa = 6.57FKK19 pKa = 10.75LVINPNKK26 pKa = 9.64FLSIGFCDD34 pKa = 4.01NPLMCCYY41 pKa = 10.28PEE43 pKa = 4.52LLPEE47 pKa = 5.49FGTVWDD53 pKa = 4.87CDD55 pKa = 3.54QSPLQIYY62 pKa = 10.1LEE64 pKa = 4.78SILGDD69 pKa = 4.18DD70 pKa = 4.34EE71 pKa = 4.37WSSTYY76 pKa = 10.23EE77 pKa = 4.27AIDD80 pKa = 3.54PVVPPMHH87 pKa = 6.63WNEE90 pKa = 3.79AGKK93 pKa = 10.0IFQPHH98 pKa = 6.94PGVLMHH104 pKa = 6.82HH105 pKa = 7.38IIGEE109 pKa = 4.26VAKK112 pKa = 10.58AWDD115 pKa = 3.79PNLPLFRR122 pKa = 11.84LEE124 pKa = 4.98ADD126 pKa = 4.41DD127 pKa = 5.31GSITAPEE134 pKa = 3.98QGTVVGGVIAEE145 pKa = 4.57PSSQMSTAADD155 pKa = 3.57MASGKK160 pKa = 10.41SVDD163 pKa = 4.22SEE165 pKa = 4.08WEE167 pKa = 3.9AFFSFHH173 pKa = 6.68TSVNWSTSEE182 pKa = 4.04TQGKK186 pKa = 9.14ILFKK190 pKa = 10.81QSLGPLLNPYY200 pKa = 10.02LEE202 pKa = 4.97HH203 pKa = 7.51LSKK206 pKa = 11.04LYY208 pKa = 10.13VAWSGSVEE216 pKa = 3.89VRR218 pKa = 11.84FSISGSGVFGGKK230 pKa = 9.18LAAIVVPPGVDD241 pKa = 4.14PIQSTSMLQYY251 pKa = 9.97PHH253 pKa = 7.23VLFDD257 pKa = 3.64ARR259 pKa = 11.84QVEE262 pKa = 4.59PVIFTIPDD270 pKa = 3.33LRR272 pKa = 11.84STLYY276 pKa = 10.94HH277 pKa = 6.94LMSDD281 pKa = 3.39TDD283 pKa = 3.85TTSLVIMVYY292 pKa = 10.82NDD294 pKa = 5.27LINPYY299 pKa = 10.63ANDD302 pKa = 4.11SNSSGCIVTVEE313 pKa = 4.42TKK315 pKa = 10.21PGSDD319 pKa = 3.78FKK321 pKa = 11.38FHH323 pKa = 6.82LLKK326 pKa = 10.8PPGSMLTHH334 pKa = 6.5GSVPSDD340 pKa = 4.83LIPKK344 pKa = 9.05TSSLWIGNRR353 pKa = 11.84FWSDD357 pKa = 2.1ITDD360 pKa = 3.77FVIRR364 pKa = 11.84PFVFQANRR372 pKa = 11.84HH373 pKa = 5.0FDD375 pKa = 3.97FNQEE379 pKa = 3.77TAGWSTPRR387 pKa = 11.84FRR389 pKa = 11.84PITVTISEE397 pKa = 4.38KK398 pKa = 10.83NGAKK402 pKa = 10.27LGVGVATDD410 pKa = 4.37FIVPGIPDD418 pKa = 4.36GWPDD422 pKa = 3.32TTIGEE427 pKa = 4.51KK428 pKa = 10.3LVPAGDD434 pKa = 3.8YY435 pKa = 11.01AITNGSGNDD444 pKa = 2.89ITTANQYY451 pKa = 10.38DD452 pKa = 3.63AADD455 pKa = 4.27IIRR458 pKa = 11.84NNTNFKK464 pKa = 10.68GMYY467 pKa = 9.16ICGSLQRR474 pKa = 11.84AWGDD478 pKa = 3.38KK479 pKa = 10.31KK480 pKa = 10.73ISNTAFITTATVEE493 pKa = 4.32GNDD496 pKa = 4.81LIPSNVIDD504 pKa = 3.57QTKK507 pKa = 10.17IAIFQDD513 pKa = 2.99NHH515 pKa = 5.82VQDD518 pKa = 4.55EE519 pKa = 4.71VQTSDD524 pKa = 4.02DD525 pKa = 3.57TLALLGYY532 pKa = 8.74TGIGEE537 pKa = 4.21EE538 pKa = 4.09AIGANRR544 pKa = 11.84EE545 pKa = 3.91RR546 pKa = 11.84VVRR549 pKa = 11.84ISTLPEE555 pKa = 3.42TGARR559 pKa = 11.84GGNHH563 pKa = 7.11PIFYY567 pKa = 10.22KK568 pKa = 10.91NSIKK572 pKa = 10.4LGYY575 pKa = 9.15VIRR578 pKa = 11.84SIDD581 pKa = 3.5VFNSQILHH589 pKa = 6.01TSRR592 pKa = 11.84QLSLNHH598 pKa = 6.17YY599 pKa = 8.3LLPPDD604 pKa = 3.46SFAVYY609 pKa = 10.34RR610 pKa = 11.84IIDD613 pKa = 3.9SNGSWFDD620 pKa = 3.28VGIDD624 pKa = 3.52FDD626 pKa = 4.01GFSFVGVSDD635 pKa = 3.54VGKK638 pKa = 11.02LEE640 pKa = 4.2FPLTASYY647 pKa = 9.91MGIQLAKK654 pKa = 9.99IRR656 pKa = 11.84LASNIRR662 pKa = 11.84STMTKK667 pKa = 10.33LL668 pKa = 3.23
MM1 pKa = 7.93CSTCANVLKK10 pKa = 10.53YY11 pKa = 10.45YY12 pKa = 10.68NWDD15 pKa = 3.24PHH17 pKa = 6.57FKK19 pKa = 10.75LVINPNKK26 pKa = 9.64FLSIGFCDD34 pKa = 4.01NPLMCCYY41 pKa = 10.28PEE43 pKa = 4.52LLPEE47 pKa = 5.49FGTVWDD53 pKa = 4.87CDD55 pKa = 3.54QSPLQIYY62 pKa = 10.1LEE64 pKa = 4.78SILGDD69 pKa = 4.18DD70 pKa = 4.34EE71 pKa = 4.37WSSTYY76 pKa = 10.23EE77 pKa = 4.27AIDD80 pKa = 3.54PVVPPMHH87 pKa = 6.63WNEE90 pKa = 3.79AGKK93 pKa = 10.0IFQPHH98 pKa = 6.94PGVLMHH104 pKa = 6.82HH105 pKa = 7.38IIGEE109 pKa = 4.26VAKK112 pKa = 10.58AWDD115 pKa = 3.79PNLPLFRR122 pKa = 11.84LEE124 pKa = 4.98ADD126 pKa = 4.41DD127 pKa = 5.31GSITAPEE134 pKa = 3.98QGTVVGGVIAEE145 pKa = 4.57PSSQMSTAADD155 pKa = 3.57MASGKK160 pKa = 10.41SVDD163 pKa = 4.22SEE165 pKa = 4.08WEE167 pKa = 3.9AFFSFHH173 pKa = 6.68TSVNWSTSEE182 pKa = 4.04TQGKK186 pKa = 9.14ILFKK190 pKa = 10.81QSLGPLLNPYY200 pKa = 10.02LEE202 pKa = 4.97HH203 pKa = 7.51LSKK206 pKa = 11.04LYY208 pKa = 10.13VAWSGSVEE216 pKa = 3.89VRR218 pKa = 11.84FSISGSGVFGGKK230 pKa = 9.18LAAIVVPPGVDD241 pKa = 4.14PIQSTSMLQYY251 pKa = 9.97PHH253 pKa = 7.23VLFDD257 pKa = 3.64ARR259 pKa = 11.84QVEE262 pKa = 4.59PVIFTIPDD270 pKa = 3.33LRR272 pKa = 11.84STLYY276 pKa = 10.94HH277 pKa = 6.94LMSDD281 pKa = 3.39TDD283 pKa = 3.85TTSLVIMVYY292 pKa = 10.82NDD294 pKa = 5.27LINPYY299 pKa = 10.63ANDD302 pKa = 4.11SNSSGCIVTVEE313 pKa = 4.42TKK315 pKa = 10.21PGSDD319 pKa = 3.78FKK321 pKa = 11.38FHH323 pKa = 6.82LLKK326 pKa = 10.8PPGSMLTHH334 pKa = 6.5GSVPSDD340 pKa = 4.83LIPKK344 pKa = 9.05TSSLWIGNRR353 pKa = 11.84FWSDD357 pKa = 2.1ITDD360 pKa = 3.77FVIRR364 pKa = 11.84PFVFQANRR372 pKa = 11.84HH373 pKa = 5.0FDD375 pKa = 3.97FNQEE379 pKa = 3.77TAGWSTPRR387 pKa = 11.84FRR389 pKa = 11.84PITVTISEE397 pKa = 4.38KK398 pKa = 10.83NGAKK402 pKa = 10.27LGVGVATDD410 pKa = 4.37FIVPGIPDD418 pKa = 4.36GWPDD422 pKa = 3.32TTIGEE427 pKa = 4.51KK428 pKa = 10.3LVPAGDD434 pKa = 3.8YY435 pKa = 11.01AITNGSGNDD444 pKa = 2.89ITTANQYY451 pKa = 10.38DD452 pKa = 3.63AADD455 pKa = 4.27IIRR458 pKa = 11.84NNTNFKK464 pKa = 10.68GMYY467 pKa = 9.16ICGSLQRR474 pKa = 11.84AWGDD478 pKa = 3.38KK479 pKa = 10.31KK480 pKa = 10.73ISNTAFITTATVEE493 pKa = 4.32GNDD496 pKa = 4.81LIPSNVIDD504 pKa = 3.57QTKK507 pKa = 10.17IAIFQDD513 pKa = 2.99NHH515 pKa = 5.82VQDD518 pKa = 4.55EE519 pKa = 4.71VQTSDD524 pKa = 4.02DD525 pKa = 3.57TLALLGYY532 pKa = 8.74TGIGEE537 pKa = 4.21EE538 pKa = 4.09AIGANRR544 pKa = 11.84EE545 pKa = 3.91RR546 pKa = 11.84VVRR549 pKa = 11.84ISTLPEE555 pKa = 3.42TGARR559 pKa = 11.84GGNHH563 pKa = 7.11PIFYY567 pKa = 10.22KK568 pKa = 10.91NSIKK572 pKa = 10.4LGYY575 pKa = 9.15VIRR578 pKa = 11.84SIDD581 pKa = 3.5VFNSQILHH589 pKa = 6.01TSRR592 pKa = 11.84QLSLNHH598 pKa = 6.17YY599 pKa = 8.3LLPPDD604 pKa = 3.46SFAVYY609 pKa = 10.34RR610 pKa = 11.84IIDD613 pKa = 3.9SNGSWFDD620 pKa = 3.28VGIDD624 pKa = 3.52FDD626 pKa = 4.01GFSFVGVSDD635 pKa = 3.54VGKK638 pKa = 11.02LEE640 pKa = 4.2FPLTASYY647 pKa = 9.91MGIQLAKK654 pKa = 9.99IRR656 pKa = 11.84LASNIRR662 pKa = 11.84STMTKK667 pKa = 10.33LL668 pKa = 3.23
Molecular weight: 73.52 kDa
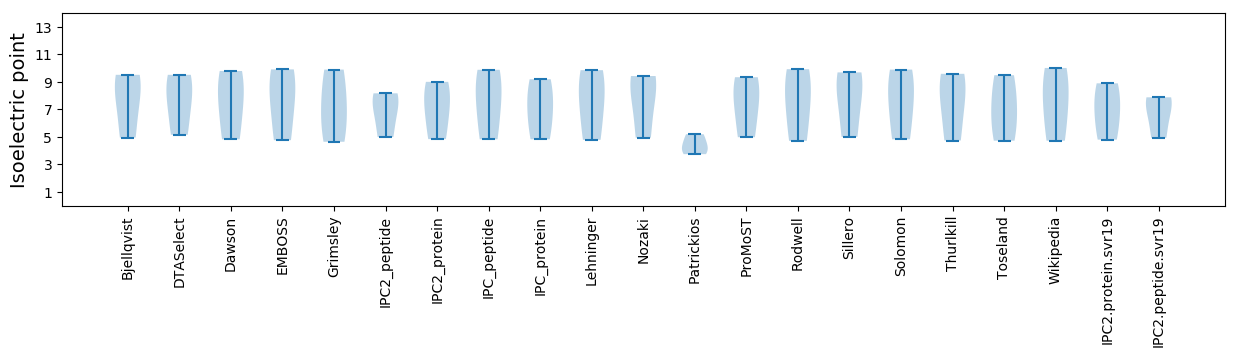
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q66916|VP2_FCVUR Protein VP2 OS=Feline calicivirus (strain Cat/United States/Urbana/1960) OX=292349 GN=ORF3 PE=1 SV=1
MM1 pKa = 7.42NSILGLIDD9 pKa = 3.47TVTNTIGKK17 pKa = 8.41AQQIEE22 pKa = 4.4LDD24 pKa = 3.79KK25 pKa = 11.3AALGQQRR32 pKa = 11.84EE33 pKa = 4.2LALQRR38 pKa = 11.84MNLDD42 pKa = 3.13RR43 pKa = 11.84QALNNQVEE51 pKa = 4.53QFNKK55 pKa = 9.93ILEE58 pKa = 4.14QRR60 pKa = 11.84VQGPLQSVRR69 pKa = 11.84LARR72 pKa = 11.84AAGFRR77 pKa = 11.84VDD79 pKa = 3.85PYY81 pKa = 11.45SYY83 pKa = 10.68TNQNFYY89 pKa = 11.26DD90 pKa = 4.02DD91 pKa = 3.8QLNAIRR97 pKa = 11.84LSYY100 pKa = 11.1RR101 pKa = 11.84NLFKK105 pKa = 11.17NN106 pKa = 3.62
MM1 pKa = 7.42NSILGLIDD9 pKa = 3.47TVTNTIGKK17 pKa = 8.41AQQIEE22 pKa = 4.4LDD24 pKa = 3.79KK25 pKa = 11.3AALGQQRR32 pKa = 11.84EE33 pKa = 4.2LALQRR38 pKa = 11.84MNLDD42 pKa = 3.13RR43 pKa = 11.84QALNNQVEE51 pKa = 4.53QFNKK55 pKa = 9.93ILEE58 pKa = 4.14QRR60 pKa = 11.84VQGPLQSVRR69 pKa = 11.84LARR72 pKa = 11.84AAGFRR77 pKa = 11.84VDD79 pKa = 3.85PYY81 pKa = 11.45SYY83 pKa = 10.68TNQNFYY89 pKa = 11.26DD90 pKa = 4.02DD91 pKa = 3.8QLNAIRR97 pKa = 11.84LSYY100 pKa = 11.1RR101 pKa = 11.84NLFKK105 pKa = 11.17NN106 pKa = 3.62
Molecular weight: 12.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2537 |
106 |
1763 |
845.7 |
93.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.095 ± 0.687 | 2.05 ± 0.642 |
5.952 ± 0.382 | 4.612 ± 0.358 |
3.981 ± 0.62 | 6.74 ± 0.567 |
2.799 ± 0.612 | 6.425 ± 0.732 |
5.676 ± 1.038 | 8.593 ± 0.87 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.128 ± 0.111 | 4.375 ± 1.273 |
5.361 ± 0.707 | 3.114 ± 1.739 |
4.533 ± 0.935 | 8.356 ± 0.849 |
6.661 ± 0.54 | 6.937 ± 0.412 |
1.498 ± 0.359 | 3.114 ± 0.125 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
