
Duck hepatitis B virus (isolate Shanghai/DHBVQCA34) (DHBV)
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Blubervirales; Hepadnaviridae; Avihepadnavirus; Duck hepatitis B virus
Average proteome isoelectric point is 8.36
Get precalculated fractions of proteins
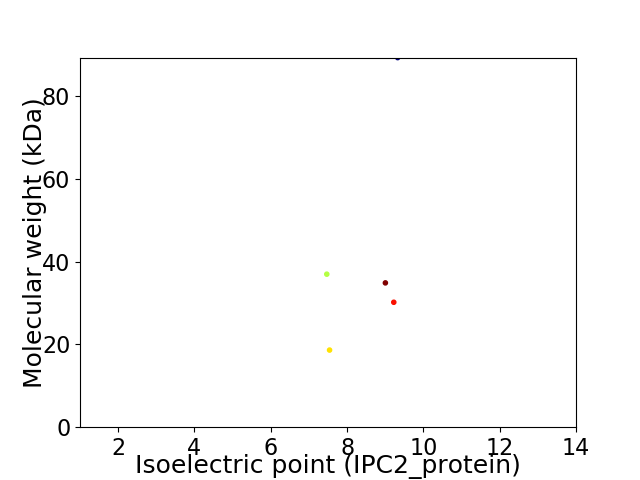
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q66406|CAPSD_DHBVQ Capsid protein OS=Duck hepatitis B virus (isolate Shanghai/DHBVQCA34) OX=644639 GN=C PE=3 SV=1
MM1 pKa = 7.15GQQPAKK7 pKa = 10.78SMDD10 pKa = 2.92VRR12 pKa = 11.84RR13 pKa = 11.84IEE15 pKa = 4.69GGEE18 pKa = 3.94LLLNQLAGRR27 pKa = 11.84MIPKK31 pKa = 8.84GTVTWSGKK39 pKa = 10.16FPTIDD44 pKa = 3.24HH45 pKa = 6.83LLDD48 pKa = 3.59HH49 pKa = 6.24VQTMEE54 pKa = 4.49EE55 pKa = 4.67VNTLQQQGAWPAGAGRR71 pKa = 11.84RR72 pKa = 11.84LGLTNPTPHH81 pKa = 6.63EE82 pKa = 4.3TPQPQWTPEE91 pKa = 3.83EE92 pKa = 4.01DD93 pKa = 3.23QKK95 pKa = 11.68ARR97 pKa = 11.84EE98 pKa = 3.95AFRR101 pKa = 11.84RR102 pKa = 11.84YY103 pKa = 8.96QEE105 pKa = 3.91EE106 pKa = 4.36RR107 pKa = 11.84PPEE110 pKa = 4.12TTTIAPTSPTPWKK123 pKa = 9.68LQPGDD128 pKa = 4.75DD129 pKa = 3.94PLLEE133 pKa = 4.35NKK135 pKa = 10.1SLLEE139 pKa = 3.89THH141 pKa = 7.35PLYY144 pKa = 10.71QNPEE148 pKa = 3.83PAVPVIKK155 pKa = 9.85TPPLKK160 pKa = 10.1KK161 pKa = 10.13KK162 pKa = 10.46KK163 pKa = 9.72MPGTFGGILAGLIGLLVSFFLLIKK187 pKa = 10.07ILEE190 pKa = 4.06ILRR193 pKa = 11.84RR194 pKa = 11.84LDD196 pKa = 3.1WWWISLSSPKK206 pKa = 10.95GKK208 pKa = 8.31MQCAFQDD215 pKa = 3.05TGAQISQHH223 pKa = 5.34YY224 pKa = 9.84VGSCPWGCPGFLWTYY239 pKa = 10.39LRR241 pKa = 11.84LFIIFLLILLVAAGLLYY258 pKa = 10.12LTDD261 pKa = 3.4NMSIILEE268 pKa = 4.07KK269 pKa = 10.68LQWEE273 pKa = 4.62SVSVLFSSISSLLPSDD289 pKa = 3.65QKK291 pKa = 11.76SLVALMFGLLLIWMTSSSATQTLVTLTQLATLSVLFYY328 pKa = 11.28KK329 pKa = 10.78NN330 pKa = 2.82
MM1 pKa = 7.15GQQPAKK7 pKa = 10.78SMDD10 pKa = 2.92VRR12 pKa = 11.84RR13 pKa = 11.84IEE15 pKa = 4.69GGEE18 pKa = 3.94LLLNQLAGRR27 pKa = 11.84MIPKK31 pKa = 8.84GTVTWSGKK39 pKa = 10.16FPTIDD44 pKa = 3.24HH45 pKa = 6.83LLDD48 pKa = 3.59HH49 pKa = 6.24VQTMEE54 pKa = 4.49EE55 pKa = 4.67VNTLQQQGAWPAGAGRR71 pKa = 11.84RR72 pKa = 11.84LGLTNPTPHH81 pKa = 6.63EE82 pKa = 4.3TPQPQWTPEE91 pKa = 3.83EE92 pKa = 4.01DD93 pKa = 3.23QKK95 pKa = 11.68ARR97 pKa = 11.84EE98 pKa = 3.95AFRR101 pKa = 11.84RR102 pKa = 11.84YY103 pKa = 8.96QEE105 pKa = 3.91EE106 pKa = 4.36RR107 pKa = 11.84PPEE110 pKa = 4.12TTTIAPTSPTPWKK123 pKa = 9.68LQPGDD128 pKa = 4.75DD129 pKa = 3.94PLLEE133 pKa = 4.35NKK135 pKa = 10.1SLLEE139 pKa = 3.89THH141 pKa = 7.35PLYY144 pKa = 10.71QNPEE148 pKa = 3.83PAVPVIKK155 pKa = 9.85TPPLKK160 pKa = 10.1KK161 pKa = 10.13KK162 pKa = 10.46KK163 pKa = 9.72MPGTFGGILAGLIGLLVSFFLLIKK187 pKa = 10.07ILEE190 pKa = 4.06ILRR193 pKa = 11.84RR194 pKa = 11.84LDD196 pKa = 3.1WWWISLSSPKK206 pKa = 10.95GKK208 pKa = 8.31MQCAFQDD215 pKa = 3.05TGAQISQHH223 pKa = 5.34YY224 pKa = 9.84VGSCPWGCPGFLWTYY239 pKa = 10.39LRR241 pKa = 11.84LFIIFLLILLVAAGLLYY258 pKa = 10.12LTDD261 pKa = 3.4NMSIILEE268 pKa = 4.07KK269 pKa = 10.68LQWEE273 pKa = 4.62SVSVLFSSISSLLPSDD289 pKa = 3.65QKK291 pKa = 11.76SLVALMFGLLLIWMTSSSATQTLVTLTQLATLSVLFYY328 pKa = 11.28KK329 pKa = 10.78NN330 pKa = 2.82
Molecular weight: 36.96 kDa
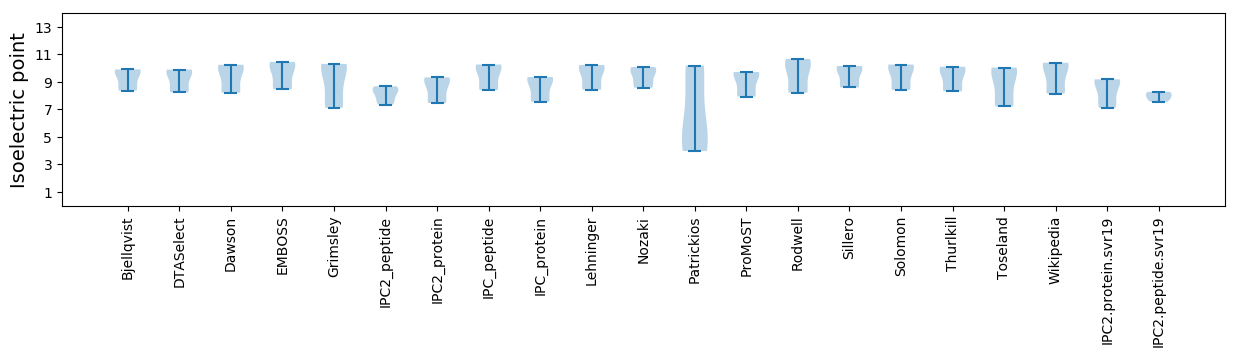
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q66405|HBSAG_DHBVQ Large envelope protein OS=Duck hepatitis B virus (isolate Shanghai/DHBVQCA34) OX=644639 GN=S PE=3 SV=1
MM1 pKa = 7.45PQPLKK6 pKa = 10.84QSLDD10 pKa = 3.17QSKK13 pKa = 10.49RR14 pKa = 11.84LRR16 pKa = 11.84EE17 pKa = 4.21AEE19 pKa = 3.75KK20 pKa = 10.54HH21 pKa = 5.62LRR23 pKa = 11.84EE24 pKa = 4.61LEE26 pKa = 4.05NLVDD30 pKa = 4.46SNLEE34 pKa = 3.97EE35 pKa = 4.51EE36 pKa = 4.37KK37 pKa = 10.94LKK39 pKa = 10.52PQLSMGEE46 pKa = 4.42DD47 pKa = 3.39VQSPGKK53 pKa = 10.64GEE55 pKa = 3.85PLHH58 pKa = 6.29PNVRR62 pKa = 11.84APLSHH67 pKa = 6.3VVRR70 pKa = 11.84AATIDD75 pKa = 3.69LPRR78 pKa = 11.84LGNKK82 pKa = 9.66LPAKK86 pKa = 9.73HH87 pKa = 6.56HH88 pKa = 6.15LGKK91 pKa = 10.87LSGLYY96 pKa = 9.38QMKK99 pKa = 9.56GCTFNPEE106 pKa = 3.58WKK108 pKa = 10.59VPDD111 pKa = 3.45ISDD114 pKa = 3.37THH116 pKa = 7.31FDD118 pKa = 3.52LQVVNEE124 pKa = 4.29CPSRR128 pKa = 11.84NWKK131 pKa = 9.93YY132 pKa = 8.85LTPAKK137 pKa = 9.91FWPKK141 pKa = 9.85SISYY145 pKa = 9.65FPVQAGVKK153 pKa = 9.77AKK155 pKa = 10.91YY156 pKa = 8.9PDD158 pKa = 3.32NVMQHH163 pKa = 5.26EE164 pKa = 4.88AIVGKK169 pKa = 9.48YY170 pKa = 9.86LNRR173 pKa = 11.84LYY175 pKa = 10.71EE176 pKa = 3.99AGILYY181 pKa = 10.13KK182 pKa = 10.39RR183 pKa = 11.84ISKK186 pKa = 10.27HH187 pKa = 5.64LVTFKK192 pKa = 10.75GKK194 pKa = 9.45PYY196 pKa = 10.15NWEE199 pKa = 3.41LQYY202 pKa = 11.07LVKK205 pKa = 10.33QHH207 pKa = 5.66QVPDD211 pKa = 3.51GTTTSKK217 pKa = 10.82INGRR221 pKa = 11.84AEE223 pKa = 3.7NRR225 pKa = 11.84RR226 pKa = 11.84RR227 pKa = 11.84RR228 pKa = 11.84APAKK232 pKa = 10.28SISRR236 pKa = 11.84PHH238 pKa = 7.13DD239 pKa = 3.41SEE241 pKa = 4.98RR242 pKa = 11.84DD243 pKa = 3.71CNMVGQISNNRR254 pKa = 11.84SSIRR258 pKa = 11.84PCANNGGGKK267 pKa = 9.71HH268 pKa = 5.9SSTTGRR274 pKa = 11.84LACWGGKK281 pKa = 8.13TIGTDD286 pKa = 3.12QSYY289 pKa = 11.12SSRR292 pKa = 11.84DD293 pKa = 3.06ASASVDD299 pKa = 3.08SRR301 pKa = 11.84GRR303 pKa = 11.84SKK305 pKa = 10.88SSRR308 pKa = 11.84GFSSISRR315 pKa = 11.84RR316 pKa = 11.84KK317 pKa = 8.34ATGNHH322 pKa = 5.05HH323 pKa = 6.24HH324 pKa = 7.01CSYY327 pKa = 9.19VTNSVEE333 pKa = 3.6ATTRR337 pKa = 11.84GRR339 pKa = 11.84STPGKK344 pKa = 10.01QVFTRR349 pKa = 11.84DD350 pKa = 3.44SSSLPEE356 pKa = 3.98SRR358 pKa = 11.84ASRR361 pKa = 11.84ACDD364 pKa = 3.47KK365 pKa = 11.04DD366 pKa = 3.78SSPQEE371 pKa = 4.3EE372 pKa = 4.04EE373 pKa = 3.9NAWYY377 pKa = 10.46LRR379 pKa = 11.84GNTSWPNRR387 pKa = 11.84ITGKK391 pKa = 10.57LFLVDD396 pKa = 3.43KK397 pKa = 10.57NSRR400 pKa = 11.84NTTEE404 pKa = 3.6ARR406 pKa = 11.84LVVDD410 pKa = 5.36FSQFSKK416 pKa = 10.78GKK418 pKa = 9.03NAMRR422 pKa = 11.84FPRR425 pKa = 11.84YY426 pKa = 8.48WSPNLSTLRR435 pKa = 11.84RR436 pKa = 11.84ILPVGMPRR444 pKa = 11.84ISLDD448 pKa = 3.21LSQAFYY454 pKa = 10.9HH455 pKa = 6.6LPLNPASSSRR465 pKa = 11.84LAVSDD470 pKa = 3.95GQHH473 pKa = 4.67VYY475 pKa = 10.42YY476 pKa = 10.43FRR478 pKa = 11.84KK479 pKa = 10.17APMGVGLSPFLLHH492 pKa = 7.37LFTTALGSEE501 pKa = 3.97IARR504 pKa = 11.84RR505 pKa = 11.84FNVWTFTYY513 pKa = 9.89MDD515 pKa = 5.26DD516 pKa = 5.74FLLCHH521 pKa = 6.51PNARR525 pKa = 11.84HH526 pKa = 6.01LNSISHH532 pKa = 6.57AVCSFLQEE540 pKa = 3.6LGIRR544 pKa = 11.84INFDD548 pKa = 2.79KK549 pKa = 8.52TTPSPVNDD557 pKa = 3.02IRR559 pKa = 11.84FLGYY563 pKa = 10.64QIDD566 pKa = 3.83QKK568 pKa = 10.99FMKK571 pKa = 9.8IEE573 pKa = 3.68EE574 pKa = 4.36SRR576 pKa = 11.84WIEE579 pKa = 3.72LRR581 pKa = 11.84TVIKK585 pKa = 10.07KK586 pKa = 10.14IKK588 pKa = 9.72IGAWYY593 pKa = 9.18DD594 pKa = 3.2WKK596 pKa = 10.91CIQRR600 pKa = 11.84FVGHH604 pKa = 6.96LNFVLPFTKK613 pKa = 10.62GNIEE617 pKa = 4.08MLKK620 pKa = 10.42PMYY623 pKa = 10.41AAITNKK629 pKa = 9.7VNFSFSSAYY638 pKa = 8.34RR639 pKa = 11.84TLLYY643 pKa = 10.37KK644 pKa = 10.16LTMGVCKK651 pKa = 10.22LAIRR655 pKa = 11.84PKK657 pKa = 10.65SSVPLPRR664 pKa = 11.84VATDD668 pKa = 3.36ATPTHH673 pKa = 6.4GAISHH678 pKa = 5.63ITGGSAVFAFSKK690 pKa = 10.69VRR692 pKa = 11.84DD693 pKa = 3.38IHH695 pKa = 7.3IQEE698 pKa = 4.6LLMVCLAKK706 pKa = 10.61IMIKK710 pKa = 9.85PRR712 pKa = 11.84CILSDD717 pKa = 3.57STFVCHH723 pKa = 7.09KK724 pKa = 10.27RR725 pKa = 11.84YY726 pKa = 8.06QTLPWHH732 pKa = 6.55FAMLAKK738 pKa = 10.32QLLSPIQLYY747 pKa = 8.05FVPSKK752 pKa = 10.83YY753 pKa = 10.6NPADD757 pKa = 3.52GPSRR761 pKa = 11.84HH762 pKa = 6.15RR763 pKa = 11.84PPDD766 pKa = 3.32WTALTYY772 pKa = 10.44TPLSKK777 pKa = 10.42AIYY780 pKa = 9.36IPHH783 pKa = 7.04RR784 pKa = 11.84LCGTT788 pKa = 3.88
MM1 pKa = 7.45PQPLKK6 pKa = 10.84QSLDD10 pKa = 3.17QSKK13 pKa = 10.49RR14 pKa = 11.84LRR16 pKa = 11.84EE17 pKa = 4.21AEE19 pKa = 3.75KK20 pKa = 10.54HH21 pKa = 5.62LRR23 pKa = 11.84EE24 pKa = 4.61LEE26 pKa = 4.05NLVDD30 pKa = 4.46SNLEE34 pKa = 3.97EE35 pKa = 4.51EE36 pKa = 4.37KK37 pKa = 10.94LKK39 pKa = 10.52PQLSMGEE46 pKa = 4.42DD47 pKa = 3.39VQSPGKK53 pKa = 10.64GEE55 pKa = 3.85PLHH58 pKa = 6.29PNVRR62 pKa = 11.84APLSHH67 pKa = 6.3VVRR70 pKa = 11.84AATIDD75 pKa = 3.69LPRR78 pKa = 11.84LGNKK82 pKa = 9.66LPAKK86 pKa = 9.73HH87 pKa = 6.56HH88 pKa = 6.15LGKK91 pKa = 10.87LSGLYY96 pKa = 9.38QMKK99 pKa = 9.56GCTFNPEE106 pKa = 3.58WKK108 pKa = 10.59VPDD111 pKa = 3.45ISDD114 pKa = 3.37THH116 pKa = 7.31FDD118 pKa = 3.52LQVVNEE124 pKa = 4.29CPSRR128 pKa = 11.84NWKK131 pKa = 9.93YY132 pKa = 8.85LTPAKK137 pKa = 9.91FWPKK141 pKa = 9.85SISYY145 pKa = 9.65FPVQAGVKK153 pKa = 9.77AKK155 pKa = 10.91YY156 pKa = 8.9PDD158 pKa = 3.32NVMQHH163 pKa = 5.26EE164 pKa = 4.88AIVGKK169 pKa = 9.48YY170 pKa = 9.86LNRR173 pKa = 11.84LYY175 pKa = 10.71EE176 pKa = 3.99AGILYY181 pKa = 10.13KK182 pKa = 10.39RR183 pKa = 11.84ISKK186 pKa = 10.27HH187 pKa = 5.64LVTFKK192 pKa = 10.75GKK194 pKa = 9.45PYY196 pKa = 10.15NWEE199 pKa = 3.41LQYY202 pKa = 11.07LVKK205 pKa = 10.33QHH207 pKa = 5.66QVPDD211 pKa = 3.51GTTTSKK217 pKa = 10.82INGRR221 pKa = 11.84AEE223 pKa = 3.7NRR225 pKa = 11.84RR226 pKa = 11.84RR227 pKa = 11.84RR228 pKa = 11.84APAKK232 pKa = 10.28SISRR236 pKa = 11.84PHH238 pKa = 7.13DD239 pKa = 3.41SEE241 pKa = 4.98RR242 pKa = 11.84DD243 pKa = 3.71CNMVGQISNNRR254 pKa = 11.84SSIRR258 pKa = 11.84PCANNGGGKK267 pKa = 9.71HH268 pKa = 5.9SSTTGRR274 pKa = 11.84LACWGGKK281 pKa = 8.13TIGTDD286 pKa = 3.12QSYY289 pKa = 11.12SSRR292 pKa = 11.84DD293 pKa = 3.06ASASVDD299 pKa = 3.08SRR301 pKa = 11.84GRR303 pKa = 11.84SKK305 pKa = 10.88SSRR308 pKa = 11.84GFSSISRR315 pKa = 11.84RR316 pKa = 11.84KK317 pKa = 8.34ATGNHH322 pKa = 5.05HH323 pKa = 6.24HH324 pKa = 7.01CSYY327 pKa = 9.19VTNSVEE333 pKa = 3.6ATTRR337 pKa = 11.84GRR339 pKa = 11.84STPGKK344 pKa = 10.01QVFTRR349 pKa = 11.84DD350 pKa = 3.44SSSLPEE356 pKa = 3.98SRR358 pKa = 11.84ASRR361 pKa = 11.84ACDD364 pKa = 3.47KK365 pKa = 11.04DD366 pKa = 3.78SSPQEE371 pKa = 4.3EE372 pKa = 4.04EE373 pKa = 3.9NAWYY377 pKa = 10.46LRR379 pKa = 11.84GNTSWPNRR387 pKa = 11.84ITGKK391 pKa = 10.57LFLVDD396 pKa = 3.43KK397 pKa = 10.57NSRR400 pKa = 11.84NTTEE404 pKa = 3.6ARR406 pKa = 11.84LVVDD410 pKa = 5.36FSQFSKK416 pKa = 10.78GKK418 pKa = 9.03NAMRR422 pKa = 11.84FPRR425 pKa = 11.84YY426 pKa = 8.48WSPNLSTLRR435 pKa = 11.84RR436 pKa = 11.84ILPVGMPRR444 pKa = 11.84ISLDD448 pKa = 3.21LSQAFYY454 pKa = 10.9HH455 pKa = 6.6LPLNPASSSRR465 pKa = 11.84LAVSDD470 pKa = 3.95GQHH473 pKa = 4.67VYY475 pKa = 10.42YY476 pKa = 10.43FRR478 pKa = 11.84KK479 pKa = 10.17APMGVGLSPFLLHH492 pKa = 7.37LFTTALGSEE501 pKa = 3.97IARR504 pKa = 11.84RR505 pKa = 11.84FNVWTFTYY513 pKa = 9.89MDD515 pKa = 5.26DD516 pKa = 5.74FLLCHH521 pKa = 6.51PNARR525 pKa = 11.84HH526 pKa = 6.01LNSISHH532 pKa = 6.57AVCSFLQEE540 pKa = 3.6LGIRR544 pKa = 11.84INFDD548 pKa = 2.79KK549 pKa = 8.52TTPSPVNDD557 pKa = 3.02IRR559 pKa = 11.84FLGYY563 pKa = 10.64QIDD566 pKa = 3.83QKK568 pKa = 10.99FMKK571 pKa = 9.8IEE573 pKa = 3.68EE574 pKa = 4.36SRR576 pKa = 11.84WIEE579 pKa = 3.72LRR581 pKa = 11.84TVIKK585 pKa = 10.07KK586 pKa = 10.14IKK588 pKa = 9.72IGAWYY593 pKa = 9.18DD594 pKa = 3.2WKK596 pKa = 10.91CIQRR600 pKa = 11.84FVGHH604 pKa = 6.96LNFVLPFTKK613 pKa = 10.62GNIEE617 pKa = 4.08MLKK620 pKa = 10.42PMYY623 pKa = 10.41AAITNKK629 pKa = 9.7VNFSFSSAYY638 pKa = 8.34RR639 pKa = 11.84TLLYY643 pKa = 10.37KK644 pKa = 10.16LTMGVCKK651 pKa = 10.22LAIRR655 pKa = 11.84PKK657 pKa = 10.65SSVPLPRR664 pKa = 11.84VATDD668 pKa = 3.36ATPTHH673 pKa = 6.4GAISHH678 pKa = 5.63ITGGSAVFAFSKK690 pKa = 10.69VRR692 pKa = 11.84DD693 pKa = 3.38IHH695 pKa = 7.3IQEE698 pKa = 4.6LLMVCLAKK706 pKa = 10.61IMIKK710 pKa = 9.85PRR712 pKa = 11.84CILSDD717 pKa = 3.57STFVCHH723 pKa = 7.09KK724 pKa = 10.27RR725 pKa = 11.84YY726 pKa = 8.06QTLPWHH732 pKa = 6.55FAMLAKK738 pKa = 10.32QLLSPIQLYY747 pKa = 8.05FVPSKK752 pKa = 10.83YY753 pKa = 10.6NPADD757 pKa = 3.52GPSRR761 pKa = 11.84HH762 pKa = 6.15RR763 pKa = 11.84PPDD766 pKa = 3.32WTALTYY772 pKa = 10.44TPLSKK777 pKa = 10.42AIYY780 pKa = 9.36IPHH783 pKa = 7.04RR784 pKa = 11.84LCGTT788 pKa = 3.88
Molecular weight: 89.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1852 |
167 |
788 |
370.4 |
41.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.479 ± 0.582 | 1.404 ± 0.261 |
4.158 ± 0.429 | 4.482 ± 0.566 |
3.672 ± 0.406 | 5.616 ± 0.468 |
2.916 ± 0.434 | 5.94 ± 0.434 |
5.832 ± 0.536 | 11.069 ± 1.56 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.89 ± 0.327 | 3.186 ± 0.686 |
6.911 ± 0.487 | 3.942 ± 0.553 |
7.127 ± 1.163 | 8.747 ± 0.592 |
6.317 ± 0.42 | 4.806 ± 0.25 |
2.43 ± 0.34 | 3.078 ± 0.3 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
