
Bacteroides sp. CAG:443
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; environmental samples
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
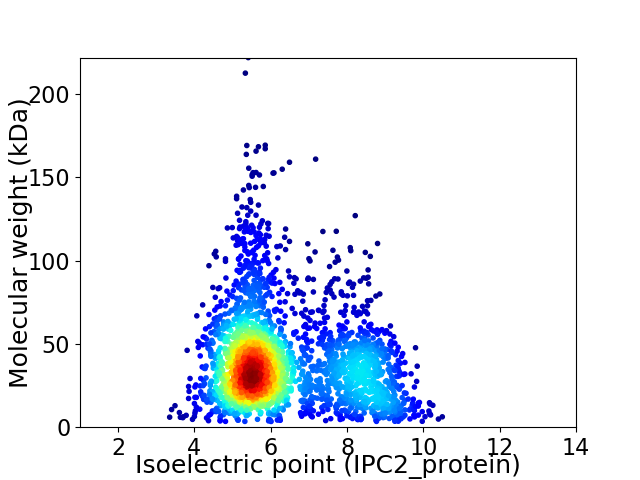
Virtual 2D-PAGE plot for 2710 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6MMB2|R6MMB2_9BACE Chaperone protein ClpB OS=Bacteroides sp. CAG:443 OX=1262739 GN=clpB PE=3 SV=1
MM1 pKa = 7.46KK2 pKa = 10.48AILKK6 pKa = 8.64YY7 pKa = 10.5IKK9 pKa = 9.9GLVMIGAIISGMVVTSCEE27 pKa = 4.16DD28 pKa = 3.34EE29 pKa = 4.09PDD31 pKa = 3.31KK32 pKa = 11.88YY33 pKa = 10.77EE34 pKa = 4.6ISDD37 pKa = 3.61GLPTVKK43 pKa = 10.14YY44 pKa = 10.82IRR46 pKa = 11.84MTDD49 pKa = 3.55PEE51 pKa = 4.47VADD54 pKa = 3.76SLITGAYY61 pKa = 6.97MANTICLVGEE71 pKa = 4.19NLRR74 pKa = 11.84SVYY77 pKa = 10.24EE78 pKa = 4.66LYY80 pKa = 11.08FNDD83 pKa = 3.99QKK85 pKa = 11.66AILNTSLITDD95 pKa = 3.27NTLIVDD101 pKa = 4.35IPSTIPSVVTNKK113 pKa = 9.91IYY115 pKa = 9.74LTNKK119 pKa = 7.81NHH121 pKa = 5.61EE122 pKa = 4.38TVTYY126 pKa = 9.83DD127 pKa = 3.63FNVLVPAPEE136 pKa = 3.89ITSMSNEE143 pKa = 3.83YY144 pKa = 10.13AQPGTVATIYY154 pKa = 10.99GDD156 pKa = 3.84YY157 pKa = 10.82FIDD160 pKa = 4.6DD161 pKa = 4.4PNVPLSIEE169 pKa = 3.89IGNVPVTEE177 pKa = 4.16ITDD180 pKa = 3.64LTRR183 pKa = 11.84NAVSFVIPDD192 pKa = 3.84GAEE195 pKa = 3.21MGYY198 pKa = 10.31INVEE202 pKa = 4.36SIYY205 pKa = 10.34GTGQSTFRR213 pKa = 11.84YY214 pKa = 8.11MEE216 pKa = 4.55RR217 pKa = 11.84DD218 pKa = 3.54GILFDD223 pKa = 5.57FDD225 pKa = 6.26DD226 pKa = 5.82DD227 pKa = 5.05GDD229 pKa = 4.12DD230 pKa = 6.49AIAKK234 pKa = 10.03DD235 pKa = 4.24NGWHH239 pKa = 6.61AATIGTLDD247 pKa = 3.88KK248 pKa = 11.16VASLDD253 pKa = 3.5GNYY256 pKa = 10.6LIFQGDD262 pKa = 4.17LDD264 pKa = 3.96NGAWNDD270 pKa = 2.9TDD272 pKa = 4.49FAFEE276 pKa = 3.81YY277 pKa = 9.83WPYY280 pKa = 11.13GDD282 pKa = 4.48EE283 pKa = 3.98ATPYY287 pKa = 11.04LNTLVDD293 pKa = 5.94FEE295 pKa = 5.28DD296 pKa = 4.84LSNLQLKK303 pKa = 10.27FEE305 pKa = 4.83VNVPDD310 pKa = 3.56AWSACAMQVLLTTNNEE326 pKa = 3.85VSGNNMNNTFFQGDD340 pKa = 3.96TFPRR344 pKa = 11.84ALWIPWQSTGSYY356 pKa = 7.07TTTGWTTVTIPLSDD370 pKa = 3.65FRR372 pKa = 11.84YY373 pKa = 8.1GASGISISPLSATDD387 pKa = 3.26ITGLTLFVWNGGVTGTACSPTICIDD412 pKa = 3.59NIRR415 pKa = 11.84VVPIYY420 pKa = 10.97
MM1 pKa = 7.46KK2 pKa = 10.48AILKK6 pKa = 8.64YY7 pKa = 10.5IKK9 pKa = 9.9GLVMIGAIISGMVVTSCEE27 pKa = 4.16DD28 pKa = 3.34EE29 pKa = 4.09PDD31 pKa = 3.31KK32 pKa = 11.88YY33 pKa = 10.77EE34 pKa = 4.6ISDD37 pKa = 3.61GLPTVKK43 pKa = 10.14YY44 pKa = 10.82IRR46 pKa = 11.84MTDD49 pKa = 3.55PEE51 pKa = 4.47VADD54 pKa = 3.76SLITGAYY61 pKa = 6.97MANTICLVGEE71 pKa = 4.19NLRR74 pKa = 11.84SVYY77 pKa = 10.24EE78 pKa = 4.66LYY80 pKa = 11.08FNDD83 pKa = 3.99QKK85 pKa = 11.66AILNTSLITDD95 pKa = 3.27NTLIVDD101 pKa = 4.35IPSTIPSVVTNKK113 pKa = 9.91IYY115 pKa = 9.74LTNKK119 pKa = 7.81NHH121 pKa = 5.61EE122 pKa = 4.38TVTYY126 pKa = 9.83DD127 pKa = 3.63FNVLVPAPEE136 pKa = 3.89ITSMSNEE143 pKa = 3.83YY144 pKa = 10.13AQPGTVATIYY154 pKa = 10.99GDD156 pKa = 3.84YY157 pKa = 10.82FIDD160 pKa = 4.6DD161 pKa = 4.4PNVPLSIEE169 pKa = 3.89IGNVPVTEE177 pKa = 4.16ITDD180 pKa = 3.64LTRR183 pKa = 11.84NAVSFVIPDD192 pKa = 3.84GAEE195 pKa = 3.21MGYY198 pKa = 10.31INVEE202 pKa = 4.36SIYY205 pKa = 10.34GTGQSTFRR213 pKa = 11.84YY214 pKa = 8.11MEE216 pKa = 4.55RR217 pKa = 11.84DD218 pKa = 3.54GILFDD223 pKa = 5.57FDD225 pKa = 6.26DD226 pKa = 5.82DD227 pKa = 5.05GDD229 pKa = 4.12DD230 pKa = 6.49AIAKK234 pKa = 10.03DD235 pKa = 4.24NGWHH239 pKa = 6.61AATIGTLDD247 pKa = 3.88KK248 pKa = 11.16VASLDD253 pKa = 3.5GNYY256 pKa = 10.6LIFQGDD262 pKa = 4.17LDD264 pKa = 3.96NGAWNDD270 pKa = 2.9TDD272 pKa = 4.49FAFEE276 pKa = 3.81YY277 pKa = 9.83WPYY280 pKa = 11.13GDD282 pKa = 4.48EE283 pKa = 3.98ATPYY287 pKa = 11.04LNTLVDD293 pKa = 5.94FEE295 pKa = 5.28DD296 pKa = 4.84LSNLQLKK303 pKa = 10.27FEE305 pKa = 4.83VNVPDD310 pKa = 3.56AWSACAMQVLLTTNNEE326 pKa = 3.85VSGNNMNNTFFQGDD340 pKa = 3.96TFPRR344 pKa = 11.84ALWIPWQSTGSYY356 pKa = 7.07TTTGWTTVTIPLSDD370 pKa = 3.65FRR372 pKa = 11.84YY373 pKa = 8.1GASGISISPLSATDD387 pKa = 3.26ITGLTLFVWNGGVTGTACSPTICIDD412 pKa = 3.59NIRR415 pKa = 11.84VVPIYY420 pKa = 10.97
Molecular weight: 46.16 kDa
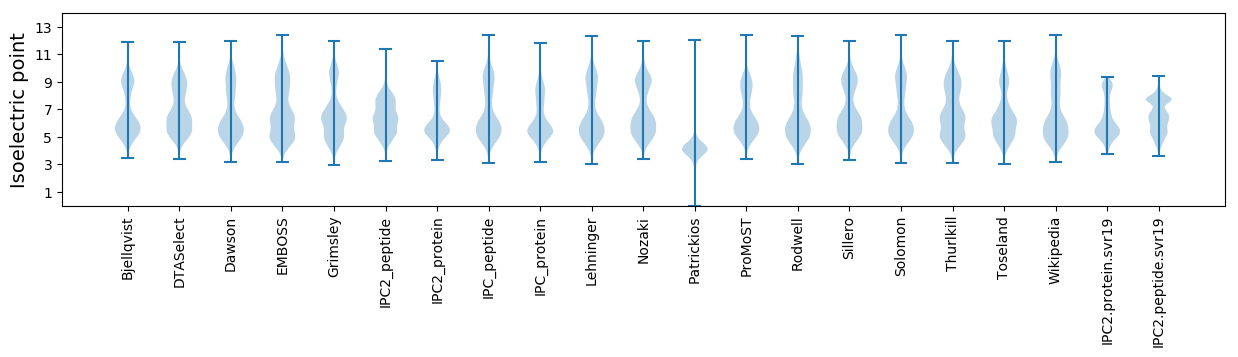
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6NKX6|R6NKX6_9BACE 3-dehydroquinate dehydratase OS=Bacteroides sp. CAG:443 OX=1262739 GN=aroQ PE=3 SV=1
MM1 pKa = 7.0ITILRR6 pKa = 11.84IFPVFCFLQEE16 pKa = 3.8LCNRR20 pKa = 11.84LSRR23 pKa = 11.84LLLEE27 pKa = 4.81KK28 pKa = 9.79AYY30 pKa = 10.81HH31 pKa = 5.51MSRR34 pKa = 11.84FQEE37 pKa = 4.23MQTEE41 pKa = 4.74SYY43 pKa = 10.83SLYY46 pKa = 10.26RR47 pKa = 11.84KK48 pKa = 9.2SRR50 pKa = 11.84KK51 pKa = 7.2QQIIFYY57 pKa = 9.37FLQQRR62 pKa = 11.84LFQQMGLPKK71 pKa = 10.07FQTPRR76 pKa = 11.84PQPYY80 pKa = 8.93PVKK83 pKa = 10.34HH84 pKa = 6.86DD85 pKa = 3.99SLPLSQHH92 pKa = 5.45NRR94 pKa = 11.84LHH96 pKa = 5.95VPSEE100 pKa = 4.06NVLLQQLFILIGTAIQRR117 pKa = 11.84QSRR120 pKa = 11.84RR121 pKa = 11.84VNN123 pKa = 3.11
MM1 pKa = 7.0ITILRR6 pKa = 11.84IFPVFCFLQEE16 pKa = 3.8LCNRR20 pKa = 11.84LSRR23 pKa = 11.84LLLEE27 pKa = 4.81KK28 pKa = 9.79AYY30 pKa = 10.81HH31 pKa = 5.51MSRR34 pKa = 11.84FQEE37 pKa = 4.23MQTEE41 pKa = 4.74SYY43 pKa = 10.83SLYY46 pKa = 10.26RR47 pKa = 11.84KK48 pKa = 9.2SRR50 pKa = 11.84KK51 pKa = 7.2QQIIFYY57 pKa = 9.37FLQQRR62 pKa = 11.84LFQQMGLPKK71 pKa = 10.07FQTPRR76 pKa = 11.84PQPYY80 pKa = 8.93PVKK83 pKa = 10.34HH84 pKa = 6.86DD85 pKa = 3.99SLPLSQHH92 pKa = 5.45NRR94 pKa = 11.84LHH96 pKa = 5.95VPSEE100 pKa = 4.06NVLLQQLFILIGTAIQRR117 pKa = 11.84QSRR120 pKa = 11.84RR121 pKa = 11.84VNN123 pKa = 3.11
Molecular weight: 14.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
996151 |
29 |
1963 |
367.6 |
41.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.121 ± 0.039 | 1.321 ± 0.017 |
5.499 ± 0.029 | 6.536 ± 0.041 |
4.488 ± 0.027 | 6.876 ± 0.039 |
1.961 ± 0.02 | 6.798 ± 0.038 |
6.468 ± 0.036 | 9.134 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.805 ± 0.019 | 5.058 ± 0.038 |
3.736 ± 0.022 | 3.611 ± 0.031 |
4.525 ± 0.032 | 6.069 ± 0.037 |
5.681 ± 0.031 | 6.513 ± 0.038 |
1.255 ± 0.018 | 4.543 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
