
Lettuce infectious yellows virus (isolate United States/92) (LIYV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Closteroviridae; Crinivirus; Lettuce infectious yellows virus
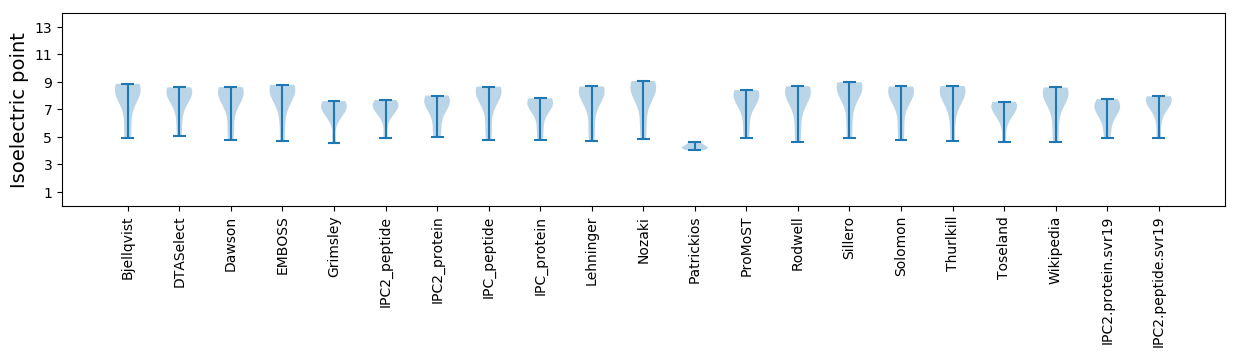
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
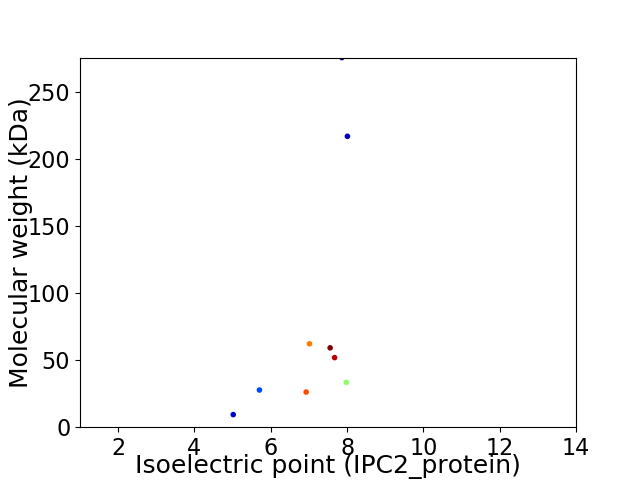
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q83044|R1A_LIYV9 Replicase polyprotein 1a OS=Lettuce infectious yellows virus (isolate United States/92) OX=651355 GN=ORF1a PE=4 SV=1
MM1 pKa = 7.49DD2 pKa = 4.51TKK4 pKa = 10.85TLIDD8 pKa = 4.14KK9 pKa = 10.8YY10 pKa = 10.84NIEE13 pKa = 4.23NFTNYY18 pKa = 9.79INFIIRR24 pKa = 11.84NHH26 pKa = 5.21QAGKK30 pKa = 10.19GNLRR34 pKa = 11.84FLVNLLKK41 pKa = 8.82TTGGSNLKK49 pKa = 10.01EE50 pKa = 3.66LDD52 pKa = 3.52INPVEE57 pKa = 4.22IEE59 pKa = 4.14NFNIDD64 pKa = 2.87IYY66 pKa = 11.36LDD68 pKa = 3.65FLEE71 pKa = 5.24FCLDD75 pKa = 3.47SKK77 pKa = 11.04FIFF80 pKa = 4.34
MM1 pKa = 7.49DD2 pKa = 4.51TKK4 pKa = 10.85TLIDD8 pKa = 4.14KK9 pKa = 10.8YY10 pKa = 10.84NIEE13 pKa = 4.23NFTNYY18 pKa = 9.79INFIIRR24 pKa = 11.84NHH26 pKa = 5.21QAGKK30 pKa = 10.19GNLRR34 pKa = 11.84FLVNLLKK41 pKa = 8.82TTGGSNLKK49 pKa = 10.01EE50 pKa = 3.66LDD52 pKa = 3.52INPVEE57 pKa = 4.22IEE59 pKa = 4.14NFNIDD64 pKa = 2.87IYY66 pKa = 11.36LDD68 pKa = 3.65FLEE71 pKa = 5.24FCLDD75 pKa = 3.47SKK77 pKa = 11.04FIFF80 pKa = 4.34
Molecular weight: 9.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q83047|MVP_LIYV9 Movement protein Hsp70h OS=Lettuce infectious yellows virus (isolate United States/92) OX=651355 PE=3 SV=1
MM1 pKa = 8.83IMMSPLYY8 pKa = 10.42ALTKK12 pKa = 10.14QCVIDD17 pKa = 3.38TAYY20 pKa = 10.86RR21 pKa = 11.84LAVPTQHH28 pKa = 6.55CAIYY32 pKa = 9.48TVACRR37 pKa = 11.84ILFLSVGFMTIVKK50 pKa = 10.11LCGFKK55 pKa = 9.87MDD57 pKa = 3.64TSSFIASIEE66 pKa = 3.92KK67 pKa = 10.49DD68 pKa = 3.31NLMDD72 pKa = 4.2CLISLVEE79 pKa = 3.81MRR81 pKa = 11.84DD82 pKa = 3.43RR83 pKa = 11.84LRR85 pKa = 11.84LCNDD89 pKa = 3.77FPILNYY95 pKa = 10.18GVNILEE101 pKa = 4.11LLIGKK106 pKa = 9.14RR107 pKa = 11.84LNKK110 pKa = 9.83INNLKK115 pKa = 8.6NCYY118 pKa = 9.57VIRR121 pKa = 11.84EE122 pKa = 4.74LITINISKK130 pKa = 9.91EE131 pKa = 4.12WVGKK135 pKa = 9.32QALKK139 pKa = 10.99VGLHH143 pKa = 6.26CFLNLSQADD152 pKa = 3.9SRR154 pKa = 11.84HH155 pKa = 4.35VKK157 pKa = 10.47YY158 pKa = 10.76LLSDD162 pKa = 3.66KK163 pKa = 11.02EE164 pKa = 4.68SLNKK168 pKa = 9.83MNFSRR173 pKa = 11.84YY174 pKa = 8.42YY175 pKa = 8.19VPKK178 pKa = 10.62VVTDD182 pKa = 4.5LYY184 pKa = 11.24LDD186 pKa = 4.97LIGVLYY192 pKa = 11.18VNTGYY197 pKa = 11.09NIDD200 pKa = 3.95LVEE203 pKa = 4.61KK204 pKa = 10.31FIFDD208 pKa = 3.39KK209 pKa = 11.62LEE211 pKa = 3.7FLVYY215 pKa = 10.57DD216 pKa = 3.73GEE218 pKa = 4.56EE219 pKa = 4.57GFKK222 pKa = 10.8SPQVEE227 pKa = 4.27YY228 pKa = 11.58NDD230 pKa = 3.02ICTVNNLKK238 pKa = 10.37PIIKK242 pKa = 9.4YY243 pKa = 9.71NRR245 pKa = 11.84WHH247 pKa = 6.6TDD249 pKa = 2.56GSIVIEE255 pKa = 4.58CGDD258 pKa = 3.97VIGKK262 pKa = 9.42GINKK266 pKa = 7.65TKK268 pKa = 10.63KK269 pKa = 9.92KK270 pKa = 10.24FAINDD275 pKa = 3.4AKK277 pKa = 11.37AEE279 pKa = 4.01FVKK282 pKa = 10.79NFKK285 pKa = 10.59AKK287 pKa = 10.46NKK289 pKa = 10.1NNEE292 pKa = 3.68
MM1 pKa = 8.83IMMSPLYY8 pKa = 10.42ALTKK12 pKa = 10.14QCVIDD17 pKa = 3.38TAYY20 pKa = 10.86RR21 pKa = 11.84LAVPTQHH28 pKa = 6.55CAIYY32 pKa = 9.48TVACRR37 pKa = 11.84ILFLSVGFMTIVKK50 pKa = 10.11LCGFKK55 pKa = 9.87MDD57 pKa = 3.64TSSFIASIEE66 pKa = 3.92KK67 pKa = 10.49DD68 pKa = 3.31NLMDD72 pKa = 4.2CLISLVEE79 pKa = 3.81MRR81 pKa = 11.84DD82 pKa = 3.43RR83 pKa = 11.84LRR85 pKa = 11.84LCNDD89 pKa = 3.77FPILNYY95 pKa = 10.18GVNILEE101 pKa = 4.11LLIGKK106 pKa = 9.14RR107 pKa = 11.84LNKK110 pKa = 9.83INNLKK115 pKa = 8.6NCYY118 pKa = 9.57VIRR121 pKa = 11.84EE122 pKa = 4.74LITINISKK130 pKa = 9.91EE131 pKa = 4.12WVGKK135 pKa = 9.32QALKK139 pKa = 10.99VGLHH143 pKa = 6.26CFLNLSQADD152 pKa = 3.9SRR154 pKa = 11.84HH155 pKa = 4.35VKK157 pKa = 10.47YY158 pKa = 10.76LLSDD162 pKa = 3.66KK163 pKa = 11.02EE164 pKa = 4.68SLNKK168 pKa = 9.83MNFSRR173 pKa = 11.84YY174 pKa = 8.42YY175 pKa = 8.19VPKK178 pKa = 10.62VVTDD182 pKa = 4.5LYY184 pKa = 11.24LDD186 pKa = 4.97LIGVLYY192 pKa = 11.18VNTGYY197 pKa = 11.09NIDD200 pKa = 3.95LVEE203 pKa = 4.61KK204 pKa = 10.31FIFDD208 pKa = 3.39KK209 pKa = 11.62LEE211 pKa = 3.7FLVYY215 pKa = 10.57DD216 pKa = 3.73GEE218 pKa = 4.56EE219 pKa = 4.57GFKK222 pKa = 10.8SPQVEE227 pKa = 4.27YY228 pKa = 11.58NDD230 pKa = 3.02ICTVNNLKK238 pKa = 10.37PIIKK242 pKa = 9.4YY243 pKa = 9.71NRR245 pKa = 11.84WHH247 pKa = 6.6TDD249 pKa = 2.56GSIVIEE255 pKa = 4.58CGDD258 pKa = 3.97VIGKK262 pKa = 9.42GINKK266 pKa = 7.65TKK268 pKa = 10.63KK269 pKa = 9.92KK270 pKa = 10.24FAINDD275 pKa = 3.4AKK277 pKa = 11.37AEE279 pKa = 4.01FVKK282 pKa = 10.79NFKK285 pKa = 10.59AKK287 pKa = 10.46NKK289 pKa = 10.1NNEE292 pKa = 3.68
Molecular weight: 33.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6620 |
80 |
2379 |
735.6 |
84.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.063 ± 0.277 | 2.13 ± 0.239 |
6.616 ± 0.353 | 5.755 ± 0.37 |
5.332 ± 0.157 | 4.562 ± 0.25 |
1.813 ± 0.207 | 7.764 ± 0.346 |
8.52 ± 0.266 | 8.867 ± 0.356 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.085 ± 0.116 | 6.737 ± 0.433 |
2.931 ± 0.182 | 2.356 ± 0.181 |
5.151 ± 0.341 | 7.689 ± 0.283 |
5.483 ± 0.185 | 6.36 ± 0.334 |
0.967 ± 0.161 | 4.819 ± 0.24 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
