
African eggplant yellowing virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus; unclassified Polerovirus
Average proteome isoelectric point is 8.02
Get precalculated fractions of proteins
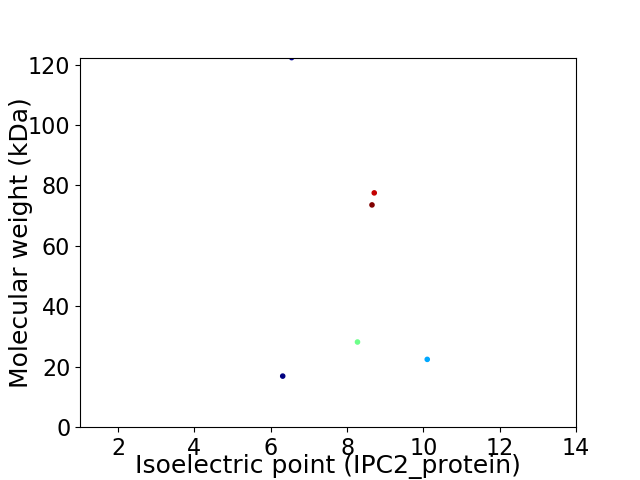
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S6PCS2|A0A1S6PCS2_9LUTE Readthrough protein OS=African eggplant yellowing virus OX=1963256 PE=3 SV=1
MM1 pKa = 7.37VKK3 pKa = 10.03ILLALFAFFSLFSLGTPSHH22 pKa = 6.8LGLPLDD28 pKa = 3.76GTLFRR33 pKa = 11.84DD34 pKa = 3.67SGNEE38 pKa = 4.02LIASPSTAEE47 pKa = 3.96LRR49 pKa = 11.84HH50 pKa = 5.85FPSINLSVLRR60 pKa = 11.84PMRR63 pKa = 11.84SPGRR67 pKa = 11.84EE68 pKa = 3.83TLDD71 pKa = 3.51SKK73 pKa = 11.74DD74 pKa = 3.49LTFSDD79 pKa = 4.93AYY81 pKa = 10.66RR82 pKa = 11.84LMWEE86 pKa = 3.92VSLRR90 pKa = 11.84DD91 pKa = 3.8TKK93 pKa = 10.9HH94 pKa = 6.26LSLEE98 pKa = 4.02ATRR101 pKa = 11.84ILQNSFASGRR111 pKa = 11.84ATLTNCLEE119 pKa = 4.28KK120 pKa = 10.96ALGALLWMIVWIWSSVIWAFGYY142 pKa = 6.2WTFYY146 pKa = 10.42MVTTFTMPVVCLALLFAFTKK166 pKa = 10.42FMVLAAEE173 pKa = 4.46KK174 pKa = 9.66MLAGWPVYY182 pKa = 10.06LGKK185 pKa = 10.82LFLRR189 pKa = 11.84LLKK192 pKa = 9.61TASTALFSKK201 pKa = 10.41RR202 pKa = 11.84NYY204 pKa = 9.99VPEE207 pKa = 4.2KK208 pKa = 9.76SVKK211 pKa = 10.29GFISVKK217 pKa = 9.9IPQSPPRR224 pKa = 11.84GSVLLIQHH232 pKa = 7.41DD233 pKa = 4.53DD234 pKa = 3.8GSHH237 pKa = 6.41AGYY240 pKa = 10.6ASCVKK245 pKa = 10.66LYY247 pKa = 10.92DD248 pKa = 3.51GTLALMTCEE257 pKa = 3.98HH258 pKa = 6.57VATGVEE264 pKa = 4.14GGRR267 pKa = 11.84VVSAKK272 pKa = 9.14TRR274 pKa = 11.84NKK276 pKa = 10.03IPLNLFTPLLISQKK290 pKa = 10.96GDD292 pKa = 3.14FALLSGPPNWEE303 pKa = 3.87SLLGCKK309 pKa = 9.97GAMFVPASQLAKK321 pKa = 10.42SKK323 pKa = 9.38MRR325 pKa = 11.84FYY327 pKa = 10.78FVEE330 pKa = 4.09NEE332 pKa = 3.47EE333 pKa = 3.88WMADD337 pKa = 3.17HH338 pKa = 6.89GEE340 pKa = 4.3VVSARR345 pKa = 11.84DD346 pKa = 3.02HH347 pKa = 6.07WFATTLCNSEE357 pKa = 4.56PGFSGTPIFNGKK369 pKa = 7.73TIIGVHH375 pKa = 6.16AGSEE379 pKa = 4.18VEE381 pKa = 4.25EE382 pKa = 4.34NSNLMSVIPPVPGLTTPQYY401 pKa = 10.89VYY403 pKa = 9.56EE404 pKa = 4.34TTAPQGRR411 pKa = 11.84LFIDD415 pKa = 3.48EE416 pKa = 4.66DD417 pKa = 4.34VSVLLKK423 pKa = 10.51SVKK426 pKa = 9.31TRR428 pKa = 11.84IPDD431 pKa = 3.04IAKK434 pKa = 9.25YY435 pKa = 10.73VSVSGKK441 pKa = 8.92NWADD445 pKa = 3.38YY446 pKa = 10.94RR447 pKa = 11.84SDD449 pKa = 4.31DD450 pKa = 4.57DD451 pKa = 5.76DD452 pKa = 6.11DD453 pKa = 4.89FFEE456 pKa = 5.12SSVTFEE462 pKa = 5.1GEE464 pKa = 4.14TKK466 pKa = 9.76PAPIAEE472 pKa = 4.45PVTPPIQFGTVPEE485 pKa = 4.32APVGKK490 pKa = 8.45QQWVPRR496 pKa = 11.84AQPSSGNRR504 pKa = 11.84EE505 pKa = 3.98RR506 pKa = 11.84QSCLPKK512 pKa = 9.71QQHH515 pKa = 6.52PSSEE519 pKa = 4.2AKK521 pKa = 9.93AHH523 pKa = 6.12SGGKK527 pKa = 9.87AKK529 pKa = 10.82AKK531 pKa = 10.23GPRR534 pKa = 11.84NQHH537 pKa = 5.06PAKK540 pKa = 9.97AARR543 pKa = 11.84IARR546 pKa = 11.84KK547 pKa = 9.02RR548 pKa = 11.84AQAARR553 pKa = 11.84AAEE556 pKa = 3.92EE557 pKa = 3.73RR558 pKa = 11.84AGNVRR563 pKa = 11.84KK564 pKa = 9.59AHH566 pKa = 6.2GEE568 pKa = 3.75NGGEE572 pKa = 4.04DD573 pKa = 3.01RR574 pKa = 11.84PIRR577 pKa = 11.84HH578 pKa = 6.19RR579 pKa = 11.84KK580 pKa = 8.43EE581 pKa = 4.03SGASSCRR588 pKa = 11.84EE589 pKa = 3.79GNEE592 pKa = 3.68EE593 pKa = 4.24AKK595 pKa = 10.5RR596 pKa = 11.84LSAQEE601 pKa = 3.37IAEE604 pKa = 4.02NFEE607 pKa = 4.02AHH609 pKa = 6.98FGGLYY614 pKa = 8.62EE615 pKa = 4.45WEE617 pKa = 4.49VSTSPQEE624 pKa = 3.66IPGFEE629 pKa = 4.03RR630 pKa = 11.84CGCLPRR636 pKa = 11.84YY637 pKa = 7.96YY638 pKa = 10.28FPKK641 pKa = 10.14QIQGSSWGAQLIEE654 pKa = 4.07EE655 pKa = 4.38HH656 pKa = 7.15PEE658 pKa = 3.94LGEE661 pKa = 4.08KK662 pKa = 10.03VAGFGWPLVGPRR674 pKa = 11.84AEE676 pKa = 4.33VTSLTLQAEE685 pKa = 4.3RR686 pKa = 11.84WLQRR690 pKa = 11.84AQSAKK695 pKa = 10.15IPSTEE700 pKa = 3.64DD701 pKa = 2.93RR702 pKa = 11.84EE703 pKa = 4.26RR704 pKa = 11.84VINKK708 pKa = 8.39TVDD711 pKa = 2.72AYY713 pKa = 10.34KK714 pKa = 10.37DD715 pKa = 3.72VKK717 pKa = 10.49TMGPTATRR725 pKa = 11.84GNKK728 pKa = 10.04LEE730 pKa = 3.9WSQFLEE736 pKa = 4.24DD737 pKa = 5.16FKK739 pKa = 11.03TAVNSLEE746 pKa = 3.99LDD748 pKa = 3.3AGIGVPYY755 pKa = 10.1ISYY758 pKa = 10.38GRR760 pKa = 11.84PTHH763 pKa = 6.43RR764 pKa = 11.84GWVEE768 pKa = 3.89DD769 pKa = 3.99PKK771 pKa = 11.09LLPVLARR778 pKa = 11.84LTFSRR783 pKa = 11.84LQKK786 pKa = 9.58MLEE789 pKa = 4.05VEE791 pKa = 4.35ADD793 pKa = 3.67LLTAEE798 pKa = 4.38QLVQEE803 pKa = 4.85GLCDD807 pKa = 4.87PIRR810 pKa = 11.84VFVKK814 pKa = 10.54RR815 pKa = 11.84EE816 pKa = 3.54PHH818 pKa = 5.83KK819 pKa = 10.47QSKK822 pKa = 10.11LDD824 pKa = 3.38EE825 pKa = 4.27GRR827 pKa = 11.84YY828 pKa = 8.96RR829 pKa = 11.84LIMSVSLVDD838 pKa = 3.45QLVARR843 pKa = 11.84VLFQNQNKK851 pKa = 9.66RR852 pKa = 11.84EE853 pKa = 3.89IALWRR858 pKa = 11.84CNPSKK863 pKa = 10.66PGFGLSTDD871 pKa = 3.63EE872 pKa = 3.99QVLEE876 pKa = 4.47FVEE879 pKa = 4.34TLAAQVEE886 pKa = 4.78VTPEE890 pKa = 3.77DD891 pKa = 5.07LVASWEE897 pKa = 4.34KK898 pKa = 10.96YY899 pKa = 9.98LVPTDD904 pKa = 3.67CSGFDD909 pKa = 3.21WSVAEE914 pKa = 4.57WMLHH918 pKa = 5.89DD919 pKa = 5.83DD920 pKa = 3.43MVVRR924 pKa = 11.84NKK926 pKa = 9.42LTLDD930 pKa = 3.76LNSTTEE936 pKa = 4.06KK937 pKa = 10.64LRR939 pKa = 11.84KK940 pKa = 7.13TWLRR944 pKa = 11.84CISNSVLCLSDD955 pKa = 3.31GTLLAQRR962 pKa = 11.84VPGVQKK968 pKa = 10.45SGSYY972 pKa = 8.25NTSSSNSRR980 pKa = 11.84IRR982 pKa = 11.84VMAAYY987 pKa = 9.63HH988 pKa = 6.86AGASWAIAMGDD999 pKa = 3.51DD1000 pKa = 3.67ALEE1003 pKa = 4.26SVNTNLEE1010 pKa = 4.32VYY1012 pKa = 10.11KK1013 pKa = 10.98SLGFKK1018 pKa = 10.72VEE1020 pKa = 3.93VSGQLEE1026 pKa = 4.4FCSHH1030 pKa = 6.9IFRR1033 pKa = 11.84APDD1036 pKa = 2.91LATPVNEE1043 pKa = 3.98NKK1045 pKa = 9.38MLYY1048 pKa = 10.08KK1049 pKa = 10.68LIYY1052 pKa = 9.23GYY1054 pKa = 10.97NVGCGNLEE1062 pKa = 4.03VVSNYY1067 pKa = 9.16IAACASVLNEE1077 pKa = 4.41LRR1079 pKa = 11.84HH1080 pKa = 6.43DD1081 pKa = 4.37PDD1083 pKa = 3.94SVALLYY1089 pKa = 10.28QWLVDD1094 pKa = 3.49PVLPQKK1100 pKa = 10.43II1101 pKa = 3.75
MM1 pKa = 7.37VKK3 pKa = 10.03ILLALFAFFSLFSLGTPSHH22 pKa = 6.8LGLPLDD28 pKa = 3.76GTLFRR33 pKa = 11.84DD34 pKa = 3.67SGNEE38 pKa = 4.02LIASPSTAEE47 pKa = 3.96LRR49 pKa = 11.84HH50 pKa = 5.85FPSINLSVLRR60 pKa = 11.84PMRR63 pKa = 11.84SPGRR67 pKa = 11.84EE68 pKa = 3.83TLDD71 pKa = 3.51SKK73 pKa = 11.74DD74 pKa = 3.49LTFSDD79 pKa = 4.93AYY81 pKa = 10.66RR82 pKa = 11.84LMWEE86 pKa = 3.92VSLRR90 pKa = 11.84DD91 pKa = 3.8TKK93 pKa = 10.9HH94 pKa = 6.26LSLEE98 pKa = 4.02ATRR101 pKa = 11.84ILQNSFASGRR111 pKa = 11.84ATLTNCLEE119 pKa = 4.28KK120 pKa = 10.96ALGALLWMIVWIWSSVIWAFGYY142 pKa = 6.2WTFYY146 pKa = 10.42MVTTFTMPVVCLALLFAFTKK166 pKa = 10.42FMVLAAEE173 pKa = 4.46KK174 pKa = 9.66MLAGWPVYY182 pKa = 10.06LGKK185 pKa = 10.82LFLRR189 pKa = 11.84LLKK192 pKa = 9.61TASTALFSKK201 pKa = 10.41RR202 pKa = 11.84NYY204 pKa = 9.99VPEE207 pKa = 4.2KK208 pKa = 9.76SVKK211 pKa = 10.29GFISVKK217 pKa = 9.9IPQSPPRR224 pKa = 11.84GSVLLIQHH232 pKa = 7.41DD233 pKa = 4.53DD234 pKa = 3.8GSHH237 pKa = 6.41AGYY240 pKa = 10.6ASCVKK245 pKa = 10.66LYY247 pKa = 10.92DD248 pKa = 3.51GTLALMTCEE257 pKa = 3.98HH258 pKa = 6.57VATGVEE264 pKa = 4.14GGRR267 pKa = 11.84VVSAKK272 pKa = 9.14TRR274 pKa = 11.84NKK276 pKa = 10.03IPLNLFTPLLISQKK290 pKa = 10.96GDD292 pKa = 3.14FALLSGPPNWEE303 pKa = 3.87SLLGCKK309 pKa = 9.97GAMFVPASQLAKK321 pKa = 10.42SKK323 pKa = 9.38MRR325 pKa = 11.84FYY327 pKa = 10.78FVEE330 pKa = 4.09NEE332 pKa = 3.47EE333 pKa = 3.88WMADD337 pKa = 3.17HH338 pKa = 6.89GEE340 pKa = 4.3VVSARR345 pKa = 11.84DD346 pKa = 3.02HH347 pKa = 6.07WFATTLCNSEE357 pKa = 4.56PGFSGTPIFNGKK369 pKa = 7.73TIIGVHH375 pKa = 6.16AGSEE379 pKa = 4.18VEE381 pKa = 4.25EE382 pKa = 4.34NSNLMSVIPPVPGLTTPQYY401 pKa = 10.89VYY403 pKa = 9.56EE404 pKa = 4.34TTAPQGRR411 pKa = 11.84LFIDD415 pKa = 3.48EE416 pKa = 4.66DD417 pKa = 4.34VSVLLKK423 pKa = 10.51SVKK426 pKa = 9.31TRR428 pKa = 11.84IPDD431 pKa = 3.04IAKK434 pKa = 9.25YY435 pKa = 10.73VSVSGKK441 pKa = 8.92NWADD445 pKa = 3.38YY446 pKa = 10.94RR447 pKa = 11.84SDD449 pKa = 4.31DD450 pKa = 4.57DD451 pKa = 5.76DD452 pKa = 6.11DD453 pKa = 4.89FFEE456 pKa = 5.12SSVTFEE462 pKa = 5.1GEE464 pKa = 4.14TKK466 pKa = 9.76PAPIAEE472 pKa = 4.45PVTPPIQFGTVPEE485 pKa = 4.32APVGKK490 pKa = 8.45QQWVPRR496 pKa = 11.84AQPSSGNRR504 pKa = 11.84EE505 pKa = 3.98RR506 pKa = 11.84QSCLPKK512 pKa = 9.71QQHH515 pKa = 6.52PSSEE519 pKa = 4.2AKK521 pKa = 9.93AHH523 pKa = 6.12SGGKK527 pKa = 9.87AKK529 pKa = 10.82AKK531 pKa = 10.23GPRR534 pKa = 11.84NQHH537 pKa = 5.06PAKK540 pKa = 9.97AARR543 pKa = 11.84IARR546 pKa = 11.84KK547 pKa = 9.02RR548 pKa = 11.84AQAARR553 pKa = 11.84AAEE556 pKa = 3.92EE557 pKa = 3.73RR558 pKa = 11.84AGNVRR563 pKa = 11.84KK564 pKa = 9.59AHH566 pKa = 6.2GEE568 pKa = 3.75NGGEE572 pKa = 4.04DD573 pKa = 3.01RR574 pKa = 11.84PIRR577 pKa = 11.84HH578 pKa = 6.19RR579 pKa = 11.84KK580 pKa = 8.43EE581 pKa = 4.03SGASSCRR588 pKa = 11.84EE589 pKa = 3.79GNEE592 pKa = 3.68EE593 pKa = 4.24AKK595 pKa = 10.5RR596 pKa = 11.84LSAQEE601 pKa = 3.37IAEE604 pKa = 4.02NFEE607 pKa = 4.02AHH609 pKa = 6.98FGGLYY614 pKa = 8.62EE615 pKa = 4.45WEE617 pKa = 4.49VSTSPQEE624 pKa = 3.66IPGFEE629 pKa = 4.03RR630 pKa = 11.84CGCLPRR636 pKa = 11.84YY637 pKa = 7.96YY638 pKa = 10.28FPKK641 pKa = 10.14QIQGSSWGAQLIEE654 pKa = 4.07EE655 pKa = 4.38HH656 pKa = 7.15PEE658 pKa = 3.94LGEE661 pKa = 4.08KK662 pKa = 10.03VAGFGWPLVGPRR674 pKa = 11.84AEE676 pKa = 4.33VTSLTLQAEE685 pKa = 4.3RR686 pKa = 11.84WLQRR690 pKa = 11.84AQSAKK695 pKa = 10.15IPSTEE700 pKa = 3.64DD701 pKa = 2.93RR702 pKa = 11.84EE703 pKa = 4.26RR704 pKa = 11.84VINKK708 pKa = 8.39TVDD711 pKa = 2.72AYY713 pKa = 10.34KK714 pKa = 10.37DD715 pKa = 3.72VKK717 pKa = 10.49TMGPTATRR725 pKa = 11.84GNKK728 pKa = 10.04LEE730 pKa = 3.9WSQFLEE736 pKa = 4.24DD737 pKa = 5.16FKK739 pKa = 11.03TAVNSLEE746 pKa = 3.99LDD748 pKa = 3.3AGIGVPYY755 pKa = 10.1ISYY758 pKa = 10.38GRR760 pKa = 11.84PTHH763 pKa = 6.43RR764 pKa = 11.84GWVEE768 pKa = 3.89DD769 pKa = 3.99PKK771 pKa = 11.09LLPVLARR778 pKa = 11.84LTFSRR783 pKa = 11.84LQKK786 pKa = 9.58MLEE789 pKa = 4.05VEE791 pKa = 4.35ADD793 pKa = 3.67LLTAEE798 pKa = 4.38QLVQEE803 pKa = 4.85GLCDD807 pKa = 4.87PIRR810 pKa = 11.84VFVKK814 pKa = 10.54RR815 pKa = 11.84EE816 pKa = 3.54PHH818 pKa = 5.83KK819 pKa = 10.47QSKK822 pKa = 10.11LDD824 pKa = 3.38EE825 pKa = 4.27GRR827 pKa = 11.84YY828 pKa = 8.96RR829 pKa = 11.84LIMSVSLVDD838 pKa = 3.45QLVARR843 pKa = 11.84VLFQNQNKK851 pKa = 9.66RR852 pKa = 11.84EE853 pKa = 3.89IALWRR858 pKa = 11.84CNPSKK863 pKa = 10.66PGFGLSTDD871 pKa = 3.63EE872 pKa = 3.99QVLEE876 pKa = 4.47FVEE879 pKa = 4.34TLAAQVEE886 pKa = 4.78VTPEE890 pKa = 3.77DD891 pKa = 5.07LVASWEE897 pKa = 4.34KK898 pKa = 10.96YY899 pKa = 9.98LVPTDD904 pKa = 3.67CSGFDD909 pKa = 3.21WSVAEE914 pKa = 4.57WMLHH918 pKa = 5.89DD919 pKa = 5.83DD920 pKa = 3.43MVVRR924 pKa = 11.84NKK926 pKa = 9.42LTLDD930 pKa = 3.76LNSTTEE936 pKa = 4.06KK937 pKa = 10.64LRR939 pKa = 11.84KK940 pKa = 7.13TWLRR944 pKa = 11.84CISNSVLCLSDD955 pKa = 3.31GTLLAQRR962 pKa = 11.84VPGVQKK968 pKa = 10.45SGSYY972 pKa = 8.25NTSSSNSRR980 pKa = 11.84IRR982 pKa = 11.84VMAAYY987 pKa = 9.63HH988 pKa = 6.86AGASWAIAMGDD999 pKa = 3.51DD1000 pKa = 3.67ALEE1003 pKa = 4.26SVNTNLEE1010 pKa = 4.32VYY1012 pKa = 10.11KK1013 pKa = 10.98SLGFKK1018 pKa = 10.72VEE1020 pKa = 3.93VSGQLEE1026 pKa = 4.4FCSHH1030 pKa = 6.9IFRR1033 pKa = 11.84APDD1036 pKa = 2.91LATPVNEE1043 pKa = 3.98NKK1045 pKa = 9.38MLYY1048 pKa = 10.08KK1049 pKa = 10.68LIYY1052 pKa = 9.23GYY1054 pKa = 10.97NVGCGNLEE1062 pKa = 4.03VVSNYY1067 pKa = 9.16IAACASVLNEE1077 pKa = 4.41LRR1079 pKa = 11.84HH1080 pKa = 6.43DD1081 pKa = 4.37PDD1083 pKa = 3.94SVALLYY1089 pKa = 10.28QWLVDD1094 pKa = 3.49PVLPQKK1100 pKa = 10.43II1101 pKa = 3.75
Molecular weight: 122.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S6PCU6|A0A1S6PCU6_9LUTE p3 OS=African eggplant yellowing virus OX=1963256 PE=4 SV=1
MM1 pKa = 6.8NTGGRR6 pKa = 11.84KK7 pKa = 8.27NVNYY11 pKa = 9.86RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84QRR17 pKa = 11.84RR18 pKa = 11.84TVRR21 pKa = 11.84PVQQMVVAVPSGQPRR36 pKa = 11.84GRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84NRR44 pKa = 11.84NRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84GVRR51 pKa = 11.84GGPRR55 pKa = 11.84SGGSSNSEE63 pKa = 3.35TFVFNKK69 pKa = 10.46DD70 pKa = 3.59SIKK73 pKa = 10.99DD74 pKa = 3.68NSAGTIKK81 pKa = 10.65FGPNLSEE88 pKa = 4.82SIALSGGVLKK98 pKa = 10.54AYY100 pKa = 10.22HH101 pKa = 6.68EE102 pKa = 4.5YY103 pKa = 10.51KK104 pKa = 9.32ITMVNIRR111 pKa = 11.84FVSEE115 pKa = 4.1SSSTAEE121 pKa = 3.6GSIAYY126 pKa = 9.39EE127 pKa = 4.08LDD129 pKa = 3.29PHH131 pKa = 7.03CKK133 pKa = 10.0LKK135 pKa = 10.89SLQSTLRR142 pKa = 11.84KK143 pKa = 9.77FPVTKK148 pKa = 10.49GGSTTFRR155 pKa = 11.84AAQINGVEE163 pKa = 4.07WHH165 pKa = 7.01DD166 pKa = 3.65TADD169 pKa = 3.53DD170 pKa = 3.93QFFLHH175 pKa = 6.22YY176 pKa = 10.23AGNGTSGKK184 pKa = 8.04TAGFLQIRR192 pKa = 11.84FTVQLHH198 pKa = 5.06NPKK201 pKa = 10.54
MM1 pKa = 6.8NTGGRR6 pKa = 11.84KK7 pKa = 8.27NVNYY11 pKa = 9.86RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84QRR17 pKa = 11.84RR18 pKa = 11.84TVRR21 pKa = 11.84PVQQMVVAVPSGQPRR36 pKa = 11.84GRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84NRR44 pKa = 11.84NRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84GVRR51 pKa = 11.84GGPRR55 pKa = 11.84SGGSSNSEE63 pKa = 3.35TFVFNKK69 pKa = 10.46DD70 pKa = 3.59SIKK73 pKa = 10.99DD74 pKa = 3.68NSAGTIKK81 pKa = 10.65FGPNLSEE88 pKa = 4.82SIALSGGVLKK98 pKa = 10.54AYY100 pKa = 10.22HH101 pKa = 6.68EE102 pKa = 4.5YY103 pKa = 10.51KK104 pKa = 9.32ITMVNIRR111 pKa = 11.84FVSEE115 pKa = 4.1SSSTAEE121 pKa = 3.6GSIAYY126 pKa = 9.39EE127 pKa = 4.08LDD129 pKa = 3.29PHH131 pKa = 7.03CKK133 pKa = 10.0LKK135 pKa = 10.89SLQSTLRR142 pKa = 11.84KK143 pKa = 9.77FPVTKK148 pKa = 10.49GGSTTFRR155 pKa = 11.84AAQINGVEE163 pKa = 4.07WHH165 pKa = 7.01DD166 pKa = 3.65TADD169 pKa = 3.53DD170 pKa = 3.93QFFLHH175 pKa = 6.22YY176 pKa = 10.23AGNGTSGKK184 pKa = 8.04TAGFLQIRR192 pKa = 11.84FTVQLHH198 pKa = 5.06NPKK201 pKa = 10.54
Molecular weight: 22.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3072 |
154 |
1101 |
512.0 |
56.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.552 ± 0.501 | 1.335 ± 0.349 |
4.46 ± 0.471 | 5.957 ± 0.496 |
4.264 ± 0.243 | 7.389 ± 0.623 |
2.116 ± 0.179 | 4.199 ± 0.394 |
5.664 ± 0.608 | 8.984 ± 1.242 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.758 ± 0.261 | 3.809 ± 0.447 |
6.283 ± 0.447 | 3.516 ± 0.199 |
6.836 ± 1.097 | 8.952 ± 0.639 |
5.762 ± 0.41 | 6.771 ± 0.454 |
1.823 ± 0.238 | 2.539 ± 0.185 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
