
Microlunatus sp. Gsoil 973
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Propionibacteriaceae; Microlunatus; unclassified Microlunatus
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
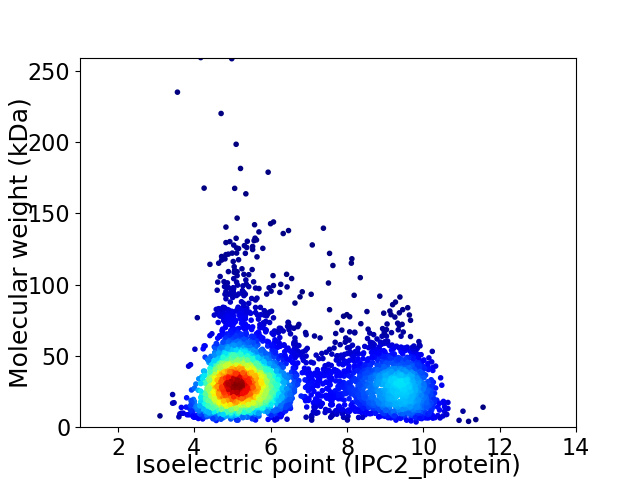
Virtual 2D-PAGE plot for 4311 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I5VNC4|A0A6I5VNC4_9ACTN DUF4097 family beta strand repeat protein OS=Microlunatus sp. Gsoil 973 OX=2672569 GN=GJV80_19935 PE=4 SV=1
MM1 pKa = 7.53EE2 pKa = 5.73SIATASFTPPPRR14 pKa = 11.84AGSYY18 pKa = 7.11YY19 pKa = 9.62WRR21 pKa = 11.84VSYY24 pKa = 11.04SGDD27 pKa = 3.57DD28 pKa = 3.51NYY30 pKa = 11.28TALPQSPCGEE40 pKa = 4.25AGEE43 pKa = 4.61TSTVTPAVPTIATQATTATLPNASVSDD70 pKa = 3.79TATVTGIPDD79 pKa = 3.43APIPTGTVTFRR90 pKa = 11.84LYY92 pKa = 10.59GASGTPVCTGDD103 pKa = 3.71PAFGPATVPLTDD115 pKa = 3.72GTATSPEE122 pKa = 4.11FSPTVAGDD130 pKa = 3.61YY131 pKa = 10.26FWLVDD136 pKa = 3.37YY137 pKa = 11.41SGDD140 pKa = 3.55ANYY143 pKa = 10.71EE144 pKa = 4.27PVTSTCDD151 pKa = 3.44DD152 pKa = 4.12PNEE155 pKa = 4.0KK156 pKa = 10.55SRR158 pKa = 11.84ITKK161 pKa = 10.25SAVLLTSSAADD172 pKa = 3.16ARR174 pKa = 11.84FGDD177 pKa = 4.58EE178 pKa = 4.66ISDD181 pKa = 3.5TGLLVGVGDD190 pKa = 3.95VAPTGTVTFTAYY202 pKa = 10.47GPTDD206 pKa = 3.31SDD208 pKa = 3.69EE209 pKa = 4.4PVCEE213 pKa = 4.24DD214 pKa = 3.89VAFAQAGTDD223 pKa = 3.66LQTTSPGTATADD235 pKa = 3.34ASFTPSDD242 pKa = 3.36PGRR245 pKa = 11.84YY246 pKa = 7.42FWRR249 pKa = 11.84ISYY252 pKa = 10.53SGDD255 pKa = 3.1SNYY258 pKa = 9.99VDD260 pKa = 3.54TVSDD264 pKa = 3.88CGALDD269 pKa = 3.45EE270 pKa = 4.58EE271 pKa = 4.99SRR273 pKa = 11.84VTVPSIDD280 pKa = 3.27IAKK283 pKa = 8.47TATPDD288 pKa = 3.32SFGSADD294 pKa = 3.93SPITYY299 pKa = 10.13NYY301 pKa = 10.44RR302 pKa = 11.84VTNTGDD308 pKa = 3.15ATLDD312 pKa = 3.49NVRR315 pKa = 11.84VTDD318 pKa = 3.63PHH320 pKa = 7.25DD321 pKa = 4.1GLSEE325 pKa = 4.03LACAPAQGSSLDD337 pKa = 3.84PGDD340 pKa = 4.64VMTCTAVYY348 pKa = 8.28ATTQADD354 pKa = 4.11LNAGSITNTATVTGTPPTGADD375 pKa = 3.35VTDD378 pKa = 3.47TDD380 pKa = 3.98QAVVTANQGPDD391 pKa = 2.83IQIAKK396 pKa = 9.15NASPGSFGAAGTPITYY412 pKa = 10.05SYY414 pKa = 11.32QVTNTGNVTLDD425 pKa = 3.7DD426 pKa = 3.87VRR428 pKa = 11.84VTDD431 pKa = 3.47PHH433 pKa = 7.47VGLSAVSCAPAQGSSLDD450 pKa = 3.84PGDD453 pKa = 4.64VMTCTAVYY461 pKa = 7.42TTTQADD467 pKa = 4.07LNAGSITNTATVTGTPPTGADD488 pKa = 3.35VTDD491 pKa = 3.47TDD493 pKa = 3.98QAVVTANQGPDD504 pKa = 2.83IQIAKK509 pKa = 9.15NASPGSFGAAGTPITYY525 pKa = 10.05SYY527 pKa = 11.32QVTNTGNVTLNNVRR541 pKa = 11.84VTDD544 pKa = 3.82PHH546 pKa = 7.44DD547 pKa = 4.11GLSDD551 pKa = 3.93LACAPNQGTSLDD563 pKa = 4.05PGDD566 pKa = 4.83VMTCTAVYY574 pKa = 7.57TTTQDD579 pKa = 3.54DD580 pKa = 4.43LNAGSITDD588 pKa = 3.61TATATGVSVAGDD600 pKa = 3.46NVLDD604 pKa = 3.85SDD606 pKa = 4.28EE607 pKa = 4.9ALVTASTAPAIDD619 pKa = 3.48IQKK622 pKa = 8.59TADD625 pKa = 3.34PTEE628 pKa = 4.56FGTAGTPITYY638 pKa = 8.82TYY640 pKa = 11.11QVTNTGNVTLDD651 pKa = 3.7DD652 pKa = 3.87VRR654 pKa = 11.84VTDD657 pKa = 3.47PHH659 pKa = 7.47VGLSAVSCAPAQGATLEE676 pKa = 5.19PGDD679 pKa = 4.98QMDD682 pKa = 3.92CTAVYY687 pKa = 7.57TTTQGDD693 pKa = 4.24LNSGSVTDD701 pKa = 3.6TATVVGSPPTGSNVTDD717 pKa = 3.41TDD719 pKa = 3.48QALITANQGPAIQIAKK735 pKa = 8.37TVAPDD740 pKa = 3.21TFGAVGTPITYY751 pKa = 10.15SYY753 pKa = 11.32QVTNTGNVTLNDD765 pKa = 3.66VQVTDD770 pKa = 3.84PHH772 pKa = 7.18DD773 pKa = 4.1GLSTISCSPSQGSSLDD789 pKa = 3.42PGSAMSCTASYY800 pKa = 11.11SATQADD806 pKa = 4.77LNIGSITDD814 pKa = 3.4TATVVGSPPTGPNVTDD830 pKa = 3.23SDD832 pKa = 4.04QALVTATTAPAIDD845 pKa = 3.64IQKK848 pKa = 10.04SAEE851 pKa = 3.94PTEE854 pKa = 4.63FSAAGTPIAYY864 pKa = 8.41TFVVTNTGNVTLDD877 pKa = 3.53NVDD880 pKa = 3.24VTDD883 pKa = 4.87PLDD886 pKa = 3.79GLTPIACDD894 pKa = 3.28PAQGVSLDD902 pKa = 4.45PGDD905 pKa = 5.11QMTCTAGYY913 pKa = 6.54TTTQADD919 pKa = 4.25LNRR922 pKa = 11.84GDD924 pKa = 3.82IANTATVVGVPPAGPNVSDD943 pKa = 5.07SDD945 pKa = 3.84QQLITANQGPEE956 pKa = 3.59IQITKK961 pKa = 9.95IADD964 pKa = 3.36PTEE967 pKa = 4.51FGSAGTPITYY977 pKa = 8.61TYY979 pKa = 11.53RR980 pKa = 11.84MINTGNVTVDD990 pKa = 3.69DD991 pKa = 4.01VRR993 pKa = 11.84ITDD996 pKa = 3.55PHH998 pKa = 7.46SGLSAISCAPAQGASLDD1015 pKa = 4.46PGDD1018 pKa = 5.41QMDD1021 pKa = 4.05CTAVYY1026 pKa = 7.57TTTQTDD1032 pKa = 3.48VNRR1035 pKa = 11.84GEE1037 pKa = 4.28IVNTATVTGTSPAGDD1052 pKa = 3.49VTDD1055 pKa = 3.76TDD1057 pKa = 4.17TALVTADD1064 pKa = 3.46QGPNITVTKK1073 pKa = 10.21IADD1076 pKa = 3.49PTEE1079 pKa = 4.42FGSAGTVINYY1089 pKa = 9.53GYY1091 pKa = 11.16LVTNNGNVTLDD1102 pKa = 3.82DD1103 pKa = 3.82VRR1105 pKa = 11.84VTDD1108 pKa = 4.03PQVGLSAVSCAPAQGASLAPGDD1130 pKa = 4.25TMSCTARR1137 pKa = 11.84YY1138 pKa = 6.14TTTQADD1144 pKa = 4.17LNAGLITNIGTVTGTPPTGPAVSDD1168 pKa = 3.57SDD1170 pKa = 4.27DD1171 pKa = 3.71AAVTANQVPAITISKK1186 pKa = 9.63IADD1189 pKa = 3.54PAEE1192 pKa = 4.44FGAAGTPITYY1202 pKa = 8.79TYY1204 pKa = 11.02RR1205 pKa = 11.84VTNTGNVTLDD1215 pKa = 3.7DD1216 pKa = 3.87VRR1218 pKa = 11.84VTDD1221 pKa = 3.47PHH1223 pKa = 7.47VGLSAVSCAPAQGATLEE1240 pKa = 5.19PGDD1243 pKa = 5.31QMDD1246 pKa = 3.87CTAAYY1251 pKa = 6.89TTTQADD1257 pKa = 4.23LNSGSVTNTATVVGQPPTGPNVTDD1281 pKa = 3.39TDD1283 pKa = 3.68QALITANQGPNIQIIKK1299 pKa = 9.21IAEE1302 pKa = 4.18PEE1304 pKa = 4.36TFGSAGTPITYY1315 pKa = 10.03SYY1317 pKa = 11.32QVTNTGNVTLDD1328 pKa = 3.7DD1329 pKa = 3.87VRR1331 pKa = 11.84VTDD1334 pKa = 3.47PHH1336 pKa = 7.47VGLSAVSCAPAQGSTLDD1353 pKa = 4.66PGDD1356 pKa = 3.52QMDD1359 pKa = 4.7CIGRR1363 pKa = 11.84YY1364 pKa = 6.44TTTQSDD1370 pKa = 4.18LNAGSVTDD1378 pKa = 3.65TATVTGEE1385 pKa = 4.24PPTGNAVTDD1394 pKa = 3.6SDD1396 pKa = 4.24QALITANQGPAITIQKK1412 pKa = 6.97TANPTEE1418 pKa = 4.48FGAAGTPITYY1428 pKa = 8.87TYY1430 pKa = 11.11QVTNTGNVTLDD1441 pKa = 3.7DD1442 pKa = 3.87VRR1444 pKa = 11.84VTDD1447 pKa = 3.47PHH1449 pKa = 7.47VGLSAVSCAPAQGATLDD1466 pKa = 4.73PGDD1469 pKa = 4.7QMDD1472 pKa = 4.05CTAVYY1477 pKa = 7.57TTTQGDD1483 pKa = 4.24LNSGSVTNTATVVGSPPTGPNVTDD1507 pKa = 3.39TDD1509 pKa = 3.68QALITANQGPAIQITKK1525 pKa = 8.65TAAPDD1530 pKa = 3.28TFGAVGTPITFTYY1543 pKa = 10.61QVTNTGNVTLDD1554 pKa = 3.7DD1555 pKa = 3.87VRR1557 pKa = 11.84VTDD1560 pKa = 3.47PHH1562 pKa = 7.47VGLSAVSCAPAQGSTLDD1579 pKa = 4.66PGDD1582 pKa = 3.52QMDD1585 pKa = 4.7CIGRR1589 pKa = 11.84YY1590 pKa = 6.44TTTQSDD1596 pKa = 4.18LNAGSVTDD1604 pKa = 3.65TATVTGEE1611 pKa = 4.28PPTGPAVTDD1620 pKa = 3.35SDD1622 pKa = 3.96QALITANQGPAITIQKK1638 pKa = 6.97TANPTEE1644 pKa = 4.48FGAAGTPITYY1654 pKa = 8.87TYY1656 pKa = 11.11QVTNTGNVTLDD1667 pKa = 3.7DD1668 pKa = 3.87VRR1670 pKa = 11.84VTDD1673 pKa = 3.47PHH1675 pKa = 7.47VGLSAVSCAPAQGATLEE1692 pKa = 5.19PGDD1695 pKa = 4.98QMDD1698 pKa = 3.92CTAVYY1703 pKa = 7.57TTTQGDD1709 pKa = 4.24LNSGSVTNTATVTGEE1724 pKa = 4.19PPTGNAVTDD1733 pKa = 3.6SDD1735 pKa = 4.24QALITANQGPAITTQKK1751 pKa = 8.61TANPTEE1757 pKa = 4.48FGAAGTPITYY1767 pKa = 8.87TYY1769 pKa = 11.11QVTNTGNVTLDD1780 pKa = 3.43NVRR1783 pKa = 11.84VTDD1786 pKa = 3.7PHH1788 pKa = 7.22AGLSAVSCAPAQGATLEE1805 pKa = 5.19PGDD1808 pKa = 5.03QMDD1811 pKa = 3.59CTGRR1815 pKa = 11.84YY1816 pKa = 5.68TTTQADD1822 pKa = 4.28LNSGSVTNTATVVGAPPTGPLVTDD1846 pKa = 3.23SDD1848 pKa = 4.45SAVVVANQAANIDD1861 pKa = 3.71ITKK1864 pKa = 10.38SAFPTEE1870 pKa = 4.72FGAAGTPINYY1880 pKa = 8.01TYY1882 pKa = 11.06QVTNNGNVTLDD1893 pKa = 3.6DD1894 pKa = 3.97VQVVDD1899 pKa = 4.29PHH1901 pKa = 7.47AGLSTIACNPPQRR1914 pKa = 11.84SSLDD1918 pKa = 3.42PGDD1921 pKa = 4.07TMQCTAEE1928 pKa = 4.16YY1929 pKa = 7.2TTTQADD1935 pKa = 4.09LNRR1938 pKa = 11.84GGIVNTATVTGTPPAGTEE1956 pKa = 4.09VTDD1959 pKa = 3.49SDD1961 pKa = 4.31SALVIANQSANINIIKK1977 pKa = 8.46TASPTEE1983 pKa = 4.18FGAAGTPITYY1993 pKa = 8.95TYY1995 pKa = 11.28LVTNNGNVTLDD2006 pKa = 3.78DD2007 pKa = 3.97VLVTDD2012 pKa = 4.13PHH2014 pKa = 7.13DD2015 pKa = 4.01GLSAITCDD2023 pKa = 3.49PAQGARR2029 pKa = 11.84LDD2031 pKa = 3.93PGEE2034 pKa = 4.91EE2035 pKa = 4.13MTCTADD2041 pKa = 3.33YY2042 pKa = 7.54TTTQDD2047 pKa = 4.51DD2048 pKa = 4.08LDD2050 pKa = 5.17AGEE2053 pKa = 5.15IINTGTTSGLPVTTDD2068 pKa = 2.92STSPRR2073 pKa = 11.84LPAALAAEE2081 pKa = 4.33RR2082 pKa = 11.84VSDD2085 pKa = 3.79SDD2087 pKa = 3.93TAIVTADD2094 pKa = 3.56QTPSIAISKK2103 pKa = 10.05SADD2106 pKa = 3.44PTSVDD2111 pKa = 3.21AAGDD2115 pKa = 3.87NIIYY2119 pKa = 10.05SYY2121 pKa = 11.2RR2122 pKa = 11.84VTNNGNVTLHH2132 pKa = 6.71DD2133 pKa = 3.78VRR2135 pKa = 11.84VTDD2138 pKa = 3.55PHH2140 pKa = 7.1QGLSDD2145 pKa = 3.76ISCEE2149 pKa = 3.85PAQGSNLEE2157 pKa = 4.22PGEE2160 pKa = 3.98QQVCTASYY2168 pKa = 8.05RR2169 pKa = 11.84TKK2171 pKa = 10.48QADD2174 pKa = 3.29IEE2176 pKa = 4.33RR2177 pKa = 11.84GSIEE2181 pKa = 4.12NVGTVAALTPNDD2193 pKa = 3.93TTVTASDD2200 pKa = 3.31TAMVSVVEE2208 pKa = 4.83KK2209 pKa = 8.07PTPPPTPTPTPSPPSPTPTCPQPPPATPSPSQPPTPPEE2247 pKa = 4.29SPTPPSTSPTPPPPCQPVATPPTSTPAPTSGPANLSGTGINPLVEE2292 pKa = 4.67TILGIGAALLLAGAVLLVVHH2312 pKa = 6.35LRR2314 pKa = 11.84RR2315 pKa = 11.84RR2316 pKa = 11.84RR2317 pKa = 11.84II2318 pKa = 3.33
MM1 pKa = 7.53EE2 pKa = 5.73SIATASFTPPPRR14 pKa = 11.84AGSYY18 pKa = 7.11YY19 pKa = 9.62WRR21 pKa = 11.84VSYY24 pKa = 11.04SGDD27 pKa = 3.57DD28 pKa = 3.51NYY30 pKa = 11.28TALPQSPCGEE40 pKa = 4.25AGEE43 pKa = 4.61TSTVTPAVPTIATQATTATLPNASVSDD70 pKa = 3.79TATVTGIPDD79 pKa = 3.43APIPTGTVTFRR90 pKa = 11.84LYY92 pKa = 10.59GASGTPVCTGDD103 pKa = 3.71PAFGPATVPLTDD115 pKa = 3.72GTATSPEE122 pKa = 4.11FSPTVAGDD130 pKa = 3.61YY131 pKa = 10.26FWLVDD136 pKa = 3.37YY137 pKa = 11.41SGDD140 pKa = 3.55ANYY143 pKa = 10.71EE144 pKa = 4.27PVTSTCDD151 pKa = 3.44DD152 pKa = 4.12PNEE155 pKa = 4.0KK156 pKa = 10.55SRR158 pKa = 11.84ITKK161 pKa = 10.25SAVLLTSSAADD172 pKa = 3.16ARR174 pKa = 11.84FGDD177 pKa = 4.58EE178 pKa = 4.66ISDD181 pKa = 3.5TGLLVGVGDD190 pKa = 3.95VAPTGTVTFTAYY202 pKa = 10.47GPTDD206 pKa = 3.31SDD208 pKa = 3.69EE209 pKa = 4.4PVCEE213 pKa = 4.24DD214 pKa = 3.89VAFAQAGTDD223 pKa = 3.66LQTTSPGTATADD235 pKa = 3.34ASFTPSDD242 pKa = 3.36PGRR245 pKa = 11.84YY246 pKa = 7.42FWRR249 pKa = 11.84ISYY252 pKa = 10.53SGDD255 pKa = 3.1SNYY258 pKa = 9.99VDD260 pKa = 3.54TVSDD264 pKa = 3.88CGALDD269 pKa = 3.45EE270 pKa = 4.58EE271 pKa = 4.99SRR273 pKa = 11.84VTVPSIDD280 pKa = 3.27IAKK283 pKa = 8.47TATPDD288 pKa = 3.32SFGSADD294 pKa = 3.93SPITYY299 pKa = 10.13NYY301 pKa = 10.44RR302 pKa = 11.84VTNTGDD308 pKa = 3.15ATLDD312 pKa = 3.49NVRR315 pKa = 11.84VTDD318 pKa = 3.63PHH320 pKa = 7.25DD321 pKa = 4.1GLSEE325 pKa = 4.03LACAPAQGSSLDD337 pKa = 3.84PGDD340 pKa = 4.64VMTCTAVYY348 pKa = 8.28ATTQADD354 pKa = 4.11LNAGSITNTATVTGTPPTGADD375 pKa = 3.35VTDD378 pKa = 3.47TDD380 pKa = 3.98QAVVTANQGPDD391 pKa = 2.83IQIAKK396 pKa = 9.15NASPGSFGAAGTPITYY412 pKa = 10.05SYY414 pKa = 11.32QVTNTGNVTLDD425 pKa = 3.7DD426 pKa = 3.87VRR428 pKa = 11.84VTDD431 pKa = 3.47PHH433 pKa = 7.47VGLSAVSCAPAQGSSLDD450 pKa = 3.84PGDD453 pKa = 4.64VMTCTAVYY461 pKa = 7.42TTTQADD467 pKa = 4.07LNAGSITNTATVTGTPPTGADD488 pKa = 3.35VTDD491 pKa = 3.47TDD493 pKa = 3.98QAVVTANQGPDD504 pKa = 2.83IQIAKK509 pKa = 9.15NASPGSFGAAGTPITYY525 pKa = 10.05SYY527 pKa = 11.32QVTNTGNVTLNNVRR541 pKa = 11.84VTDD544 pKa = 3.82PHH546 pKa = 7.44DD547 pKa = 4.11GLSDD551 pKa = 3.93LACAPNQGTSLDD563 pKa = 4.05PGDD566 pKa = 4.83VMTCTAVYY574 pKa = 7.57TTTQDD579 pKa = 3.54DD580 pKa = 4.43LNAGSITDD588 pKa = 3.61TATATGVSVAGDD600 pKa = 3.46NVLDD604 pKa = 3.85SDD606 pKa = 4.28EE607 pKa = 4.9ALVTASTAPAIDD619 pKa = 3.48IQKK622 pKa = 8.59TADD625 pKa = 3.34PTEE628 pKa = 4.56FGTAGTPITYY638 pKa = 8.82TYY640 pKa = 11.11QVTNTGNVTLDD651 pKa = 3.7DD652 pKa = 3.87VRR654 pKa = 11.84VTDD657 pKa = 3.47PHH659 pKa = 7.47VGLSAVSCAPAQGATLEE676 pKa = 5.19PGDD679 pKa = 4.98QMDD682 pKa = 3.92CTAVYY687 pKa = 7.57TTTQGDD693 pKa = 4.24LNSGSVTDD701 pKa = 3.6TATVVGSPPTGSNVTDD717 pKa = 3.41TDD719 pKa = 3.48QALITANQGPAIQIAKK735 pKa = 8.37TVAPDD740 pKa = 3.21TFGAVGTPITYY751 pKa = 10.15SYY753 pKa = 11.32QVTNTGNVTLNDD765 pKa = 3.66VQVTDD770 pKa = 3.84PHH772 pKa = 7.18DD773 pKa = 4.1GLSTISCSPSQGSSLDD789 pKa = 3.42PGSAMSCTASYY800 pKa = 11.11SATQADD806 pKa = 4.77LNIGSITDD814 pKa = 3.4TATVVGSPPTGPNVTDD830 pKa = 3.23SDD832 pKa = 4.04QALVTATTAPAIDD845 pKa = 3.64IQKK848 pKa = 10.04SAEE851 pKa = 3.94PTEE854 pKa = 4.63FSAAGTPIAYY864 pKa = 8.41TFVVTNTGNVTLDD877 pKa = 3.53NVDD880 pKa = 3.24VTDD883 pKa = 4.87PLDD886 pKa = 3.79GLTPIACDD894 pKa = 3.28PAQGVSLDD902 pKa = 4.45PGDD905 pKa = 5.11QMTCTAGYY913 pKa = 6.54TTTQADD919 pKa = 4.25LNRR922 pKa = 11.84GDD924 pKa = 3.82IANTATVVGVPPAGPNVSDD943 pKa = 5.07SDD945 pKa = 3.84QQLITANQGPEE956 pKa = 3.59IQITKK961 pKa = 9.95IADD964 pKa = 3.36PTEE967 pKa = 4.51FGSAGTPITYY977 pKa = 8.61TYY979 pKa = 11.53RR980 pKa = 11.84MINTGNVTVDD990 pKa = 3.69DD991 pKa = 4.01VRR993 pKa = 11.84ITDD996 pKa = 3.55PHH998 pKa = 7.46SGLSAISCAPAQGASLDD1015 pKa = 4.46PGDD1018 pKa = 5.41QMDD1021 pKa = 4.05CTAVYY1026 pKa = 7.57TTTQTDD1032 pKa = 3.48VNRR1035 pKa = 11.84GEE1037 pKa = 4.28IVNTATVTGTSPAGDD1052 pKa = 3.49VTDD1055 pKa = 3.76TDD1057 pKa = 4.17TALVTADD1064 pKa = 3.46QGPNITVTKK1073 pKa = 10.21IADD1076 pKa = 3.49PTEE1079 pKa = 4.42FGSAGTVINYY1089 pKa = 9.53GYY1091 pKa = 11.16LVTNNGNVTLDD1102 pKa = 3.82DD1103 pKa = 3.82VRR1105 pKa = 11.84VTDD1108 pKa = 4.03PQVGLSAVSCAPAQGASLAPGDD1130 pKa = 4.25TMSCTARR1137 pKa = 11.84YY1138 pKa = 6.14TTTQADD1144 pKa = 4.17LNAGLITNIGTVTGTPPTGPAVSDD1168 pKa = 3.57SDD1170 pKa = 4.27DD1171 pKa = 3.71AAVTANQVPAITISKK1186 pKa = 9.63IADD1189 pKa = 3.54PAEE1192 pKa = 4.44FGAAGTPITYY1202 pKa = 8.79TYY1204 pKa = 11.02RR1205 pKa = 11.84VTNTGNVTLDD1215 pKa = 3.7DD1216 pKa = 3.87VRR1218 pKa = 11.84VTDD1221 pKa = 3.47PHH1223 pKa = 7.47VGLSAVSCAPAQGATLEE1240 pKa = 5.19PGDD1243 pKa = 5.31QMDD1246 pKa = 3.87CTAAYY1251 pKa = 6.89TTTQADD1257 pKa = 4.23LNSGSVTNTATVVGQPPTGPNVTDD1281 pKa = 3.39TDD1283 pKa = 3.68QALITANQGPNIQIIKK1299 pKa = 9.21IAEE1302 pKa = 4.18PEE1304 pKa = 4.36TFGSAGTPITYY1315 pKa = 10.03SYY1317 pKa = 11.32QVTNTGNVTLDD1328 pKa = 3.7DD1329 pKa = 3.87VRR1331 pKa = 11.84VTDD1334 pKa = 3.47PHH1336 pKa = 7.47VGLSAVSCAPAQGSTLDD1353 pKa = 4.66PGDD1356 pKa = 3.52QMDD1359 pKa = 4.7CIGRR1363 pKa = 11.84YY1364 pKa = 6.44TTTQSDD1370 pKa = 4.18LNAGSVTDD1378 pKa = 3.65TATVTGEE1385 pKa = 4.24PPTGNAVTDD1394 pKa = 3.6SDD1396 pKa = 4.24QALITANQGPAITIQKK1412 pKa = 6.97TANPTEE1418 pKa = 4.48FGAAGTPITYY1428 pKa = 8.87TYY1430 pKa = 11.11QVTNTGNVTLDD1441 pKa = 3.7DD1442 pKa = 3.87VRR1444 pKa = 11.84VTDD1447 pKa = 3.47PHH1449 pKa = 7.47VGLSAVSCAPAQGATLDD1466 pKa = 4.73PGDD1469 pKa = 4.7QMDD1472 pKa = 4.05CTAVYY1477 pKa = 7.57TTTQGDD1483 pKa = 4.24LNSGSVTNTATVVGSPPTGPNVTDD1507 pKa = 3.39TDD1509 pKa = 3.68QALITANQGPAIQITKK1525 pKa = 8.65TAAPDD1530 pKa = 3.28TFGAVGTPITFTYY1543 pKa = 10.61QVTNTGNVTLDD1554 pKa = 3.7DD1555 pKa = 3.87VRR1557 pKa = 11.84VTDD1560 pKa = 3.47PHH1562 pKa = 7.47VGLSAVSCAPAQGSTLDD1579 pKa = 4.66PGDD1582 pKa = 3.52QMDD1585 pKa = 4.7CIGRR1589 pKa = 11.84YY1590 pKa = 6.44TTTQSDD1596 pKa = 4.18LNAGSVTDD1604 pKa = 3.65TATVTGEE1611 pKa = 4.28PPTGPAVTDD1620 pKa = 3.35SDD1622 pKa = 3.96QALITANQGPAITIQKK1638 pKa = 6.97TANPTEE1644 pKa = 4.48FGAAGTPITYY1654 pKa = 8.87TYY1656 pKa = 11.11QVTNTGNVTLDD1667 pKa = 3.7DD1668 pKa = 3.87VRR1670 pKa = 11.84VTDD1673 pKa = 3.47PHH1675 pKa = 7.47VGLSAVSCAPAQGATLEE1692 pKa = 5.19PGDD1695 pKa = 4.98QMDD1698 pKa = 3.92CTAVYY1703 pKa = 7.57TTTQGDD1709 pKa = 4.24LNSGSVTNTATVTGEE1724 pKa = 4.19PPTGNAVTDD1733 pKa = 3.6SDD1735 pKa = 4.24QALITANQGPAITTQKK1751 pKa = 8.61TANPTEE1757 pKa = 4.48FGAAGTPITYY1767 pKa = 8.87TYY1769 pKa = 11.11QVTNTGNVTLDD1780 pKa = 3.43NVRR1783 pKa = 11.84VTDD1786 pKa = 3.7PHH1788 pKa = 7.22AGLSAVSCAPAQGATLEE1805 pKa = 5.19PGDD1808 pKa = 5.03QMDD1811 pKa = 3.59CTGRR1815 pKa = 11.84YY1816 pKa = 5.68TTTQADD1822 pKa = 4.28LNSGSVTNTATVVGAPPTGPLVTDD1846 pKa = 3.23SDD1848 pKa = 4.45SAVVVANQAANIDD1861 pKa = 3.71ITKK1864 pKa = 10.38SAFPTEE1870 pKa = 4.72FGAAGTPINYY1880 pKa = 8.01TYY1882 pKa = 11.06QVTNNGNVTLDD1893 pKa = 3.6DD1894 pKa = 3.97VQVVDD1899 pKa = 4.29PHH1901 pKa = 7.47AGLSTIACNPPQRR1914 pKa = 11.84SSLDD1918 pKa = 3.42PGDD1921 pKa = 4.07TMQCTAEE1928 pKa = 4.16YY1929 pKa = 7.2TTTQADD1935 pKa = 4.09LNRR1938 pKa = 11.84GGIVNTATVTGTPPAGTEE1956 pKa = 4.09VTDD1959 pKa = 3.49SDD1961 pKa = 4.31SALVIANQSANINIIKK1977 pKa = 8.46TASPTEE1983 pKa = 4.18FGAAGTPITYY1993 pKa = 8.95TYY1995 pKa = 11.28LVTNNGNVTLDD2006 pKa = 3.78DD2007 pKa = 3.97VLVTDD2012 pKa = 4.13PHH2014 pKa = 7.13DD2015 pKa = 4.01GLSAITCDD2023 pKa = 3.49PAQGARR2029 pKa = 11.84LDD2031 pKa = 3.93PGEE2034 pKa = 4.91EE2035 pKa = 4.13MTCTADD2041 pKa = 3.33YY2042 pKa = 7.54TTTQDD2047 pKa = 4.51DD2048 pKa = 4.08LDD2050 pKa = 5.17AGEE2053 pKa = 5.15IINTGTTSGLPVTTDD2068 pKa = 2.92STSPRR2073 pKa = 11.84LPAALAAEE2081 pKa = 4.33RR2082 pKa = 11.84VSDD2085 pKa = 3.79SDD2087 pKa = 3.93TAIVTADD2094 pKa = 3.56QTPSIAISKK2103 pKa = 10.05SADD2106 pKa = 3.44PTSVDD2111 pKa = 3.21AAGDD2115 pKa = 3.87NIIYY2119 pKa = 10.05SYY2121 pKa = 11.2RR2122 pKa = 11.84VTNNGNVTLHH2132 pKa = 6.71DD2133 pKa = 3.78VRR2135 pKa = 11.84VTDD2138 pKa = 3.55PHH2140 pKa = 7.1QGLSDD2145 pKa = 3.76ISCEE2149 pKa = 3.85PAQGSNLEE2157 pKa = 4.22PGEE2160 pKa = 3.98QQVCTASYY2168 pKa = 8.05RR2169 pKa = 11.84TKK2171 pKa = 10.48QADD2174 pKa = 3.29IEE2176 pKa = 4.33RR2177 pKa = 11.84GSIEE2181 pKa = 4.12NVGTVAALTPNDD2193 pKa = 3.93TTVTASDD2200 pKa = 3.31TAMVSVVEE2208 pKa = 4.83KK2209 pKa = 8.07PTPPPTPTPTPSPPSPTPTCPQPPPATPSPSQPPTPPEE2247 pKa = 4.29SPTPPSTSPTPPPPCQPVATPPTSTPAPTSGPANLSGTGINPLVEE2292 pKa = 4.67TILGIGAALLLAGAVLLVVHH2312 pKa = 6.35LRR2314 pKa = 11.84RR2315 pKa = 11.84RR2316 pKa = 11.84RR2317 pKa = 11.84II2318 pKa = 3.33
Molecular weight: 234.87 kDa
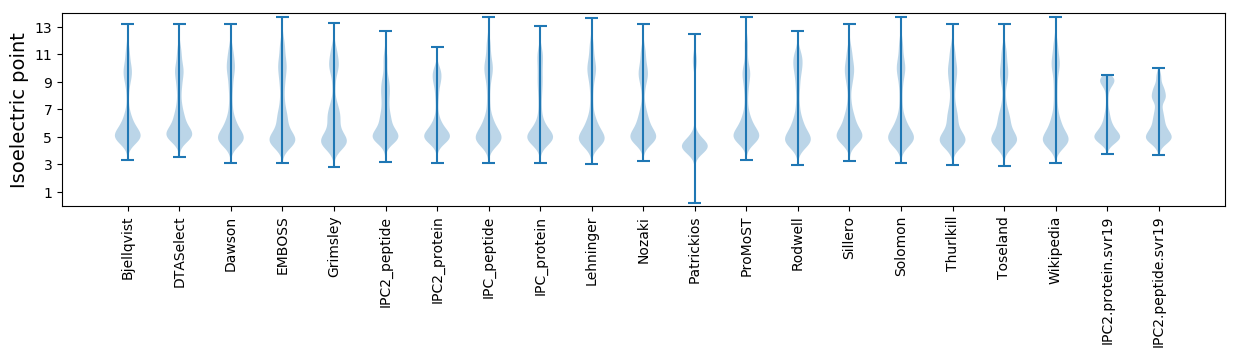
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I5VMQ4|A0A6I5VMQ4_9ACTN Zinc-binding dehydrogenase OS=Microlunatus sp. Gsoil 973 OX=2672569 GN=GJV80_16285 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SRR15 pKa = 11.84THH17 pKa = 6.24GFRR20 pKa = 11.84HH21 pKa = 6.29RR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AVLAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.68GRR40 pKa = 11.84ARR42 pKa = 11.84LAGG45 pKa = 3.63
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SRR15 pKa = 11.84THH17 pKa = 6.24GFRR20 pKa = 11.84HH21 pKa = 6.29RR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AVLAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.68GRR40 pKa = 11.84ARR42 pKa = 11.84LAGG45 pKa = 3.63
Molecular weight: 5.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1389878 |
33 |
2504 |
322.4 |
34.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.169 ± 0.051 | 0.734 ± 0.01 |
6.445 ± 0.039 | 5.109 ± 0.039 |
2.917 ± 0.023 | 9.069 ± 0.029 |
2.161 ± 0.017 | 4.685 ± 0.025 |
2.006 ± 0.03 | 10.049 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.865 ± 0.015 | 2.12 ± 0.021 |
5.783 ± 0.03 | 3.223 ± 0.019 |
7.985 ± 0.048 | 5.625 ± 0.029 |
6.013 ± 0.035 | 8.309 ± 0.037 |
1.569 ± 0.015 | 2.165 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
