
Erethizon dorsatum papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Sigmapapillomavirus; Sigmapapillomavirus 1
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
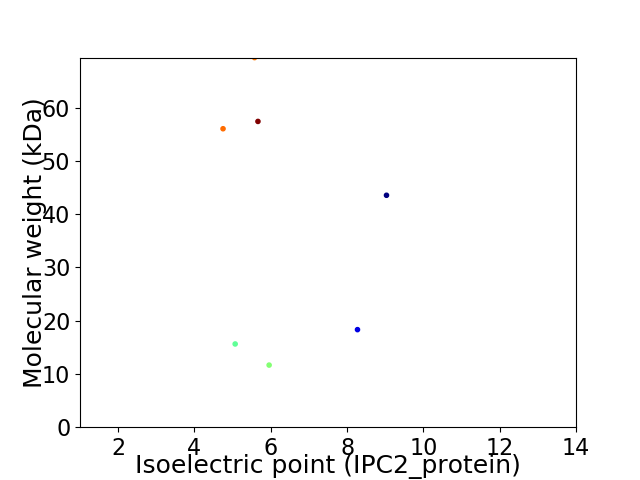
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q5IRF2|Q5IRF2_9PAPI E4 OS=Erethizon dorsatum papillomavirus 1 OX=291590 GN=E4 PE=4 SV=1
MM1 pKa = 6.98VVRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84IKK8 pKa = 10.27RR9 pKa = 11.84ANPTQLYY16 pKa = 6.6ATCKK20 pKa = 8.99ITGTCPPDD28 pKa = 4.08IINKK32 pKa = 9.51IEE34 pKa = 4.17GNTVADD40 pKa = 5.78KK41 pKa = 10.44ILKK44 pKa = 8.61WGSTGVYY51 pKa = 10.09LGGLGIGTGRR61 pKa = 11.84GGGGVSLGGGSIGSARR77 pKa = 11.84PSVPAGSIGPGDD89 pKa = 4.78IITVDD94 pKa = 3.5AEE96 pKa = 4.29AGAVQPPVEE105 pKa = 4.63PPPTTSISGSARR117 pKa = 11.84RR118 pKa = 11.84PFGEE122 pKa = 4.18GFGRR126 pKa = 11.84QPLDD130 pKa = 3.94PIGIPRR136 pKa = 11.84PRR138 pKa = 11.84PSLPRR143 pKa = 11.84PTLDD147 pKa = 3.29TVPTDD152 pKa = 3.41VAVLEE157 pKa = 5.21GPHH160 pKa = 6.0TPFGAGGDD168 pKa = 3.56IAVLEE173 pKa = 4.55VPTPEE178 pKa = 3.89VGRR181 pKa = 11.84SVTARR186 pKa = 11.84TQYY189 pKa = 11.05NNPAFEE195 pKa = 4.19VSIHH199 pKa = 6.33SEE201 pKa = 4.07LPSAEE206 pKa = 4.19TSSVDD211 pKa = 3.1HH212 pKa = 6.69VYY214 pKa = 11.02VGTPSSGSFVGQDD227 pKa = 3.34VQFEE231 pKa = 4.62LPVLQAEE238 pKa = 4.38YY239 pKa = 10.61AILDD243 pKa = 4.03GQAEE247 pKa = 4.34TSFIEE252 pKa = 4.11HH253 pKa = 6.77GSYY256 pKa = 10.82SEE258 pKa = 4.5VPLTSTPDD266 pKa = 3.39PVLPTTEE273 pKa = 5.07AIASIRR279 pKa = 11.84PYY281 pKa = 10.56GRR283 pKa = 11.84QFGQVRR289 pKa = 11.84VGDD292 pKa = 3.76AAFLSRR298 pKa = 11.84PAVLFQAEE306 pKa = 4.22NPAYY310 pKa = 10.38DD311 pKa = 4.08PDD313 pKa = 3.85VSILFEE319 pKa = 5.32RR320 pKa = 11.84DD321 pKa = 3.15VADD324 pKa = 5.36LEE326 pKa = 4.29RR327 pKa = 11.84LPDD330 pKa = 4.14PFQDD334 pKa = 2.81IQYY337 pKa = 10.51LSRR340 pKa = 11.84PYY342 pKa = 8.98YY343 pKa = 9.17QRR345 pKa = 11.84RR346 pKa = 11.84ATGLRR351 pKa = 11.84LSRR354 pKa = 11.84IGRR357 pKa = 11.84RR358 pKa = 11.84EE359 pKa = 3.86GIIQTRR365 pKa = 11.84SGTRR369 pKa = 11.84VGSSHH374 pKa = 6.76HH375 pKa = 6.04FFYY378 pKa = 10.66DD379 pKa = 3.0ISDD382 pKa = 3.5ITPLEE387 pKa = 4.53EE388 pKa = 5.05IEE390 pKa = 4.0MDD392 pKa = 4.14SIGEE396 pKa = 4.02HH397 pKa = 5.9SLGVSTAAEE406 pKa = 4.0QPIEE410 pKa = 4.34VYY412 pKa = 10.71SEE414 pKa = 4.3ADD416 pKa = 3.35LLDD419 pKa = 3.78EE420 pKa = 4.43MEE422 pKa = 5.0EE423 pKa = 4.57GDD425 pKa = 3.75TLARR429 pKa = 11.84SSQLLLSFASVGEE442 pKa = 4.1EE443 pKa = 4.5GEE445 pKa = 5.13DD446 pKa = 3.23ITIQLPIDD454 pKa = 3.77LLSIEE459 pKa = 4.51KK460 pKa = 9.98PSTAFYY466 pKa = 10.26PDD468 pKa = 2.91TGVNIAYY475 pKa = 7.48TPTKK479 pKa = 8.94ITTPSLPWVPIDD491 pKa = 3.79DD492 pKa = 4.52TLDD495 pKa = 3.16TFQNYY500 pKa = 9.19NDD502 pKa = 4.19YY503 pKa = 11.01DD504 pKa = 3.98LHH506 pKa = 6.73PSLLPRR512 pKa = 11.84RR513 pKa = 11.84KK514 pKa = 9.31RR515 pKa = 11.84KK516 pKa = 9.59RR517 pKa = 11.84RR518 pKa = 11.84ILL520 pKa = 3.67
MM1 pKa = 6.98VVRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84IKK8 pKa = 10.27RR9 pKa = 11.84ANPTQLYY16 pKa = 6.6ATCKK20 pKa = 8.99ITGTCPPDD28 pKa = 4.08IINKK32 pKa = 9.51IEE34 pKa = 4.17GNTVADD40 pKa = 5.78KK41 pKa = 10.44ILKK44 pKa = 8.61WGSTGVYY51 pKa = 10.09LGGLGIGTGRR61 pKa = 11.84GGGGVSLGGGSIGSARR77 pKa = 11.84PSVPAGSIGPGDD89 pKa = 4.78IITVDD94 pKa = 3.5AEE96 pKa = 4.29AGAVQPPVEE105 pKa = 4.63PPPTTSISGSARR117 pKa = 11.84RR118 pKa = 11.84PFGEE122 pKa = 4.18GFGRR126 pKa = 11.84QPLDD130 pKa = 3.94PIGIPRR136 pKa = 11.84PRR138 pKa = 11.84PSLPRR143 pKa = 11.84PTLDD147 pKa = 3.29TVPTDD152 pKa = 3.41VAVLEE157 pKa = 5.21GPHH160 pKa = 6.0TPFGAGGDD168 pKa = 3.56IAVLEE173 pKa = 4.55VPTPEE178 pKa = 3.89VGRR181 pKa = 11.84SVTARR186 pKa = 11.84TQYY189 pKa = 11.05NNPAFEE195 pKa = 4.19VSIHH199 pKa = 6.33SEE201 pKa = 4.07LPSAEE206 pKa = 4.19TSSVDD211 pKa = 3.1HH212 pKa = 6.69VYY214 pKa = 11.02VGTPSSGSFVGQDD227 pKa = 3.34VQFEE231 pKa = 4.62LPVLQAEE238 pKa = 4.38YY239 pKa = 10.61AILDD243 pKa = 4.03GQAEE247 pKa = 4.34TSFIEE252 pKa = 4.11HH253 pKa = 6.77GSYY256 pKa = 10.82SEE258 pKa = 4.5VPLTSTPDD266 pKa = 3.39PVLPTTEE273 pKa = 5.07AIASIRR279 pKa = 11.84PYY281 pKa = 10.56GRR283 pKa = 11.84QFGQVRR289 pKa = 11.84VGDD292 pKa = 3.76AAFLSRR298 pKa = 11.84PAVLFQAEE306 pKa = 4.22NPAYY310 pKa = 10.38DD311 pKa = 4.08PDD313 pKa = 3.85VSILFEE319 pKa = 5.32RR320 pKa = 11.84DD321 pKa = 3.15VADD324 pKa = 5.36LEE326 pKa = 4.29RR327 pKa = 11.84LPDD330 pKa = 4.14PFQDD334 pKa = 2.81IQYY337 pKa = 10.51LSRR340 pKa = 11.84PYY342 pKa = 8.98YY343 pKa = 9.17QRR345 pKa = 11.84RR346 pKa = 11.84ATGLRR351 pKa = 11.84LSRR354 pKa = 11.84IGRR357 pKa = 11.84RR358 pKa = 11.84EE359 pKa = 3.86GIIQTRR365 pKa = 11.84SGTRR369 pKa = 11.84VGSSHH374 pKa = 6.76HH375 pKa = 6.04FFYY378 pKa = 10.66DD379 pKa = 3.0ISDD382 pKa = 3.5ITPLEE387 pKa = 4.53EE388 pKa = 5.05IEE390 pKa = 4.0MDD392 pKa = 4.14SIGEE396 pKa = 4.02HH397 pKa = 5.9SLGVSTAAEE406 pKa = 4.0QPIEE410 pKa = 4.34VYY412 pKa = 10.71SEE414 pKa = 4.3ADD416 pKa = 3.35LLDD419 pKa = 3.78EE420 pKa = 4.43MEE422 pKa = 5.0EE423 pKa = 4.57GDD425 pKa = 3.75TLARR429 pKa = 11.84SSQLLLSFASVGEE442 pKa = 4.1EE443 pKa = 4.5GEE445 pKa = 5.13DD446 pKa = 3.23ITIQLPIDD454 pKa = 3.77LLSIEE459 pKa = 4.51KK460 pKa = 9.98PSTAFYY466 pKa = 10.26PDD468 pKa = 2.91TGVNIAYY475 pKa = 7.48TPTKK479 pKa = 8.94ITTPSLPWVPIDD491 pKa = 3.79DD492 pKa = 4.52TLDD495 pKa = 3.16TFQNYY500 pKa = 9.19NDD502 pKa = 4.19YY503 pKa = 11.01DD504 pKa = 3.98LHH506 pKa = 6.73PSLLPRR512 pKa = 11.84RR513 pKa = 11.84KK514 pKa = 9.31RR515 pKa = 11.84KK516 pKa = 9.59RR517 pKa = 11.84RR518 pKa = 11.84ILL520 pKa = 3.67
Molecular weight: 56.04 kDa
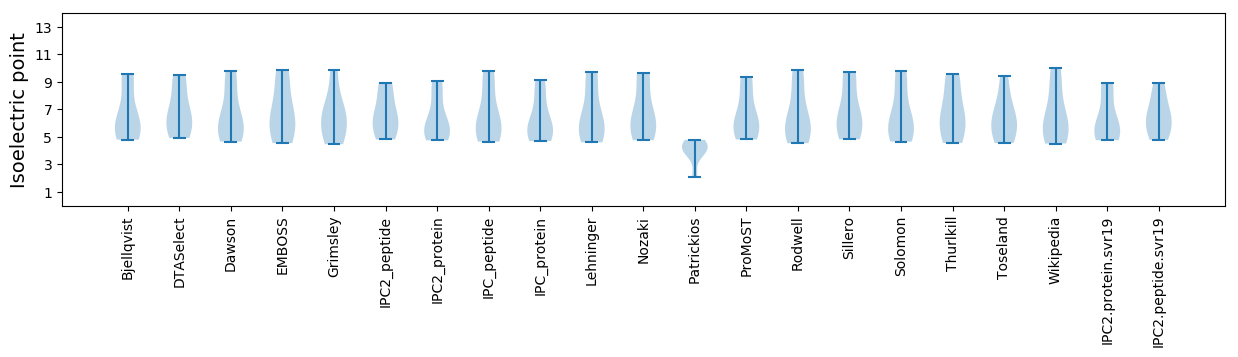
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5IRF4|Q5IRF4_9PAPI Replication protein E1 OS=Erethizon dorsatum papillomavirus 1 OX=291590 GN=E1 PE=3 SV=1
MM1 pKa = 7.57EE2 pKa = 5.79RR3 pKa = 11.84LQHH6 pKa = 6.5RR7 pKa = 11.84LDD9 pKa = 3.62YY10 pKa = 9.35TQEE13 pKa = 4.59KK14 pKa = 10.39ILTLYY19 pKa = 10.52EE20 pKa = 4.53KK21 pKa = 10.67DD22 pKa = 3.39STDD25 pKa = 3.46INDD28 pKa = 5.39HH29 pKa = 6.17ISLWMAVRR37 pKa = 11.84HH38 pKa = 5.58EE39 pKa = 4.17NVLFHH44 pKa = 6.02TMRR47 pKa = 11.84KK48 pKa = 8.63RR49 pKa = 11.84GIVRR53 pKa = 11.84VMGRR57 pKa = 11.84SIPSLQASEE66 pKa = 5.1KK67 pKa = 8.86SAKK70 pKa = 8.76HH71 pKa = 5.95AIMMQLQLEE80 pKa = 4.54DD81 pKa = 4.07LAKK84 pKa = 10.41SPYY87 pKa = 10.27GLEE90 pKa = 3.89PWSLSEE96 pKa = 4.23TTLEE100 pKa = 4.55RR101 pKa = 11.84YY102 pKa = 7.74LAPPRR107 pKa = 11.84EE108 pKa = 4.21TFKK111 pKa = 11.14KK112 pKa = 10.46GGVHH116 pKa = 6.38VNVTFDD122 pKa = 3.68NEE124 pKa = 4.05EE125 pKa = 4.33SNSAEE130 pKa = 3.9YY131 pKa = 11.35VMWTYY136 pKa = 10.92IYY138 pKa = 10.11VTDD141 pKa = 5.31EE142 pKa = 3.86NGDD145 pKa = 3.18WHH147 pKa = 5.97KK148 pKa = 9.35TQGGVDD154 pKa = 3.21RR155 pKa = 11.84TGIYY159 pKa = 10.29YY160 pKa = 9.95IGVDD164 pKa = 3.8GSHH167 pKa = 6.9IYY169 pKa = 10.26YY170 pKa = 10.5VYY172 pKa = 10.83FEE174 pKa = 5.5DD175 pKa = 4.9EE176 pKa = 4.41AAQHH180 pKa = 6.09SRR182 pKa = 11.84TGQYY186 pKa = 8.66TVNSDD191 pKa = 3.17QDD193 pKa = 3.49VSDD196 pKa = 3.99PVINTDD202 pKa = 2.79SSRR205 pKa = 11.84NRR207 pKa = 11.84EE208 pKa = 4.21TGEE211 pKa = 3.56EE212 pKa = 4.18TPRR215 pKa = 11.84RR216 pKa = 11.84RR217 pKa = 11.84GTYY220 pKa = 9.8LRR222 pKa = 11.84RR223 pKa = 11.84SSPAAPARR231 pKa = 11.84PTPPQRR237 pKa = 11.84SRR239 pKa = 11.84QRR241 pKa = 11.84KK242 pKa = 8.41RR243 pKa = 11.84SPAGPTTNTTPEE255 pKa = 4.01TRR257 pKa = 11.84PTLLSARR264 pKa = 11.84NRR266 pKa = 11.84RR267 pKa = 11.84SRR269 pKa = 11.84SGRR272 pKa = 11.84SPSATGGRR280 pKa = 11.84HH281 pKa = 4.07QARR284 pKa = 11.84HH285 pKa = 5.74RR286 pKa = 11.84SSAEE290 pKa = 3.76RR291 pKa = 11.84PGPTPQFYY299 pKa = 10.89LIGCRR304 pKa = 11.84GPVNTLRR311 pKa = 11.84CYY313 pKa = 10.39RR314 pKa = 11.84FKK316 pKa = 10.92LKK318 pKa = 10.24KK319 pKa = 10.1RR320 pKa = 11.84PNPDD324 pKa = 2.08ITYY327 pKa = 10.38ISTTFNWTAPQGTDD341 pKa = 2.42RR342 pKa = 11.84CGSGRR347 pKa = 11.84FLIAFTSKK355 pKa = 7.11EE356 pKa = 4.14HH357 pKa = 7.04RR358 pKa = 11.84DD359 pKa = 3.59EE360 pKa = 5.22FYY362 pKa = 11.05VAARR366 pKa = 11.84PPPNIGLFKK375 pKa = 11.04GEE377 pKa = 4.78CEE379 pKa = 4.42TII381 pKa = 3.47
MM1 pKa = 7.57EE2 pKa = 5.79RR3 pKa = 11.84LQHH6 pKa = 6.5RR7 pKa = 11.84LDD9 pKa = 3.62YY10 pKa = 9.35TQEE13 pKa = 4.59KK14 pKa = 10.39ILTLYY19 pKa = 10.52EE20 pKa = 4.53KK21 pKa = 10.67DD22 pKa = 3.39STDD25 pKa = 3.46INDD28 pKa = 5.39HH29 pKa = 6.17ISLWMAVRR37 pKa = 11.84HH38 pKa = 5.58EE39 pKa = 4.17NVLFHH44 pKa = 6.02TMRR47 pKa = 11.84KK48 pKa = 8.63RR49 pKa = 11.84GIVRR53 pKa = 11.84VMGRR57 pKa = 11.84SIPSLQASEE66 pKa = 5.1KK67 pKa = 8.86SAKK70 pKa = 8.76HH71 pKa = 5.95AIMMQLQLEE80 pKa = 4.54DD81 pKa = 4.07LAKK84 pKa = 10.41SPYY87 pKa = 10.27GLEE90 pKa = 3.89PWSLSEE96 pKa = 4.23TTLEE100 pKa = 4.55RR101 pKa = 11.84YY102 pKa = 7.74LAPPRR107 pKa = 11.84EE108 pKa = 4.21TFKK111 pKa = 11.14KK112 pKa = 10.46GGVHH116 pKa = 6.38VNVTFDD122 pKa = 3.68NEE124 pKa = 4.05EE125 pKa = 4.33SNSAEE130 pKa = 3.9YY131 pKa = 11.35VMWTYY136 pKa = 10.92IYY138 pKa = 10.11VTDD141 pKa = 5.31EE142 pKa = 3.86NGDD145 pKa = 3.18WHH147 pKa = 5.97KK148 pKa = 9.35TQGGVDD154 pKa = 3.21RR155 pKa = 11.84TGIYY159 pKa = 10.29YY160 pKa = 9.95IGVDD164 pKa = 3.8GSHH167 pKa = 6.9IYY169 pKa = 10.26YY170 pKa = 10.5VYY172 pKa = 10.83FEE174 pKa = 5.5DD175 pKa = 4.9EE176 pKa = 4.41AAQHH180 pKa = 6.09SRR182 pKa = 11.84TGQYY186 pKa = 8.66TVNSDD191 pKa = 3.17QDD193 pKa = 3.49VSDD196 pKa = 3.99PVINTDD202 pKa = 2.79SSRR205 pKa = 11.84NRR207 pKa = 11.84EE208 pKa = 4.21TGEE211 pKa = 3.56EE212 pKa = 4.18TPRR215 pKa = 11.84RR216 pKa = 11.84RR217 pKa = 11.84GTYY220 pKa = 9.8LRR222 pKa = 11.84RR223 pKa = 11.84SSPAAPARR231 pKa = 11.84PTPPQRR237 pKa = 11.84SRR239 pKa = 11.84QRR241 pKa = 11.84KK242 pKa = 8.41RR243 pKa = 11.84SPAGPTTNTTPEE255 pKa = 4.01TRR257 pKa = 11.84PTLLSARR264 pKa = 11.84NRR266 pKa = 11.84RR267 pKa = 11.84SRR269 pKa = 11.84SGRR272 pKa = 11.84SPSATGGRR280 pKa = 11.84HH281 pKa = 4.07QARR284 pKa = 11.84HH285 pKa = 5.74RR286 pKa = 11.84SSAEE290 pKa = 3.76RR291 pKa = 11.84PGPTPQFYY299 pKa = 10.89LIGCRR304 pKa = 11.84GPVNTLRR311 pKa = 11.84CYY313 pKa = 10.39RR314 pKa = 11.84FKK316 pKa = 10.92LKK318 pKa = 10.24KK319 pKa = 10.1RR320 pKa = 11.84PNPDD324 pKa = 2.08ITYY327 pKa = 10.38ISTTFNWTAPQGTDD341 pKa = 2.42RR342 pKa = 11.84CGSGRR347 pKa = 11.84FLIAFTSKK355 pKa = 7.11EE356 pKa = 4.14HH357 pKa = 7.04RR358 pKa = 11.84DD359 pKa = 3.59EE360 pKa = 5.22FYY362 pKa = 11.05VAARR366 pKa = 11.84PPPNIGLFKK375 pKa = 11.04GEE377 pKa = 4.78CEE379 pKa = 4.42TII381 pKa = 3.47
Molecular weight: 43.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2416 |
107 |
604 |
345.1 |
38.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.919 ± 0.433 | 2.401 ± 0.912 |
6.333 ± 0.613 | 6.126 ± 0.39 |
3.891 ± 0.514 | 6.788 ± 0.87 |
2.608 ± 0.379 | 4.76 ± 0.729 |
4.139 ± 0.752 | 8.982 ± 1.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.821 ± 0.437 | 3.394 ± 0.534 |
6.705 ± 1.069 | 4.429 ± 0.333 |
7.285 ± 0.856 | 6.664 ± 0.67 |
7.119 ± 0.514 | 5.96 ± 0.619 |
1.118 ± 0.19 | 3.56 ± 0.329 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
