
Hubei narna-like virus 25
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KI82|A0A1L3KI82_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 25 OX=1922956 PE=4 SV=1
MM1 pKa = 7.64KK2 pKa = 7.96MTKK5 pKa = 10.37FNMFKK10 pKa = 9.54VTLKK14 pKa = 10.24VVTWMFQGFNLKK26 pKa = 9.7MSGRR30 pKa = 11.84MSDD33 pKa = 3.87YY34 pKa = 11.26LDD36 pKa = 5.73LILSLDD42 pKa = 3.35RR43 pKa = 11.84LLYY46 pKa = 10.6NRR48 pKa = 11.84GALEE52 pKa = 3.71TLKK55 pKa = 10.69YY56 pKa = 10.47IKK58 pKa = 9.74GTSTAYY64 pKa = 10.3LAYY67 pKa = 10.7LSGKK71 pKa = 7.64PMRR74 pKa = 11.84VKK76 pKa = 10.62GVSTTKK82 pKa = 10.76DD83 pKa = 3.82GIPVILGDD91 pKa = 4.19FVPKK95 pKa = 9.81IRR97 pKa = 11.84KK98 pKa = 9.3GPTPAMLQLINTILFCTSALKK119 pKa = 10.77LGSTPDD125 pKa = 4.1FSPMINPPKK134 pKa = 10.11SDD136 pKa = 4.77PINMGEE142 pKa = 4.28FVDD145 pKa = 5.58DD146 pKa = 4.63FWRR149 pKa = 11.84DD150 pKa = 3.31MGYY153 pKa = 10.08HH154 pKa = 5.52RR155 pKa = 11.84QARR158 pKa = 11.84TNPKK162 pKa = 9.63SLNFRR167 pKa = 11.84KK168 pKa = 9.29FHH170 pKa = 6.31LTTKK174 pKa = 10.19SGPNGTANALWTSLNDD190 pKa = 3.66LKK192 pKa = 11.24ALSEE196 pKa = 4.1QQLTDD201 pKa = 2.87IGIIGGEE208 pKa = 4.12TLRR211 pKa = 11.84SKK213 pKa = 10.51MSTLKK218 pKa = 10.74EE219 pKa = 4.12GLDD222 pKa = 3.66KK223 pKa = 11.37VPEE226 pKa = 4.05LSYY229 pKa = 11.39FMDD232 pKa = 3.98TSAGVYY238 pKa = 6.95TSRR241 pKa = 11.84MTSFPDD247 pKa = 3.06KK248 pKa = 9.45EE249 pKa = 4.23LKK251 pKa = 10.43VRR253 pKa = 11.84VIAVGDD259 pKa = 3.97YY260 pKa = 10.13FSQAALKK267 pKa = 9.53PLHH270 pKa = 7.32DD271 pKa = 3.79YY272 pKa = 11.0LFRR275 pKa = 11.84VLKK278 pKa = 10.23KK279 pKa = 10.22IPQDD283 pKa = 3.34CTFNQGGFWDD293 pKa = 4.38KK294 pKa = 11.11IKK296 pKa = 10.8DD297 pKa = 3.55QEE299 pKa = 4.54YY300 pKa = 9.82YY301 pKa = 11.37ASIDD305 pKa = 3.31LTAATDD311 pKa = 3.6SFPISTMSQVLLGSLPKK328 pKa = 10.33SYY330 pKa = 11.12VDD332 pKa = 2.73AWHH335 pKa = 7.01RR336 pKa = 11.84LMVGTPFIYY345 pKa = 10.12EE346 pKa = 4.12GQEE349 pKa = 3.34MNYY352 pKa = 10.83AVGNPMGFYY361 pKa = 10.68SSWASFAVAHH371 pKa = 6.5HH372 pKa = 5.41YY373 pKa = 9.52VVYY376 pKa = 9.96YY377 pKa = 9.81CCKK380 pKa = 10.41KK381 pKa = 10.84LGIPWKK387 pKa = 7.7TLKK390 pKa = 10.82YY391 pKa = 10.26CLLGDD396 pKa = 5.19DD397 pKa = 5.68IVMCDD402 pKa = 3.57PKK404 pKa = 11.13VAALYY409 pKa = 10.64KK410 pKa = 9.44EE411 pKa = 4.68TISGLGVEE419 pKa = 5.03FSEE422 pKa = 4.83EE423 pKa = 3.79KK424 pKa = 10.4TITSPHH430 pKa = 5.77LVEE433 pKa = 4.07FAKK436 pKa = 10.7RR437 pKa = 11.84LIYY440 pKa = 10.42KK441 pKa = 7.7GTEE444 pKa = 3.65MSPFPISALGEE455 pKa = 4.19LANKK459 pKa = 10.31YY460 pKa = 10.41YY461 pKa = 11.2LLTDD465 pKa = 4.19LFLEE469 pKa = 4.73LEE471 pKa = 4.37SKK473 pKa = 10.6GWVSLGGAPSAVEE486 pKa = 4.16SFHH489 pKa = 7.83KK490 pKa = 10.53IVFNWSTVRR499 pKa = 11.84ARR501 pKa = 11.84EE502 pKa = 4.04IEE504 pKa = 4.49VKK506 pKa = 10.73SSVCEE511 pKa = 3.75RR512 pKa = 11.84VMKK515 pKa = 9.47MARR518 pKa = 11.84GAEE521 pKa = 4.09DD522 pKa = 3.85AGTLLTEE529 pKa = 4.69CFSKK533 pKa = 10.77LGYY536 pKa = 9.88PFVLSSFVASNVLEE550 pKa = 4.09NMVVEE555 pKa = 4.94NFANSNPASHH565 pKa = 6.63NPEE568 pKa = 3.67SDD570 pKa = 3.39KK571 pKa = 11.04KK572 pKa = 11.03SKK574 pKa = 10.57SPCVGLGYY582 pKa = 10.25LAEE585 pKa = 4.65NLTCLLTGLDD595 pKa = 4.37EE596 pKa = 4.67EE597 pKa = 4.91SCSLGFDD604 pKa = 3.51LMMALPHH611 pKa = 6.3LNAYY615 pKa = 9.56GQMSEE620 pKa = 5.0LYY622 pKa = 11.09VNINKK627 pKa = 9.84KK628 pKa = 10.07ARR630 pKa = 11.84QISTTNGAEE639 pKa = 3.83WPLLFKK645 pKa = 9.71TMCIPWDD652 pKa = 3.53DD653 pKa = 4.76TIFTMSSSHH662 pKa = 7.14LIAKK666 pKa = 9.41GSSSVVKK673 pKa = 10.39DD674 pKa = 3.62LQNSAEE680 pKa = 4.43LLSFYY685 pKa = 10.49PPEE688 pKa = 3.96EE689 pKa = 4.18LLRR692 pKa = 11.84EE693 pKa = 4.31DD694 pKa = 4.05PNFLKK699 pKa = 10.93SLL701 pKa = 3.77
MM1 pKa = 7.64KK2 pKa = 7.96MTKK5 pKa = 10.37FNMFKK10 pKa = 9.54VTLKK14 pKa = 10.24VVTWMFQGFNLKK26 pKa = 9.7MSGRR30 pKa = 11.84MSDD33 pKa = 3.87YY34 pKa = 11.26LDD36 pKa = 5.73LILSLDD42 pKa = 3.35RR43 pKa = 11.84LLYY46 pKa = 10.6NRR48 pKa = 11.84GALEE52 pKa = 3.71TLKK55 pKa = 10.69YY56 pKa = 10.47IKK58 pKa = 9.74GTSTAYY64 pKa = 10.3LAYY67 pKa = 10.7LSGKK71 pKa = 7.64PMRR74 pKa = 11.84VKK76 pKa = 10.62GVSTTKK82 pKa = 10.76DD83 pKa = 3.82GIPVILGDD91 pKa = 4.19FVPKK95 pKa = 9.81IRR97 pKa = 11.84KK98 pKa = 9.3GPTPAMLQLINTILFCTSALKK119 pKa = 10.77LGSTPDD125 pKa = 4.1FSPMINPPKK134 pKa = 10.11SDD136 pKa = 4.77PINMGEE142 pKa = 4.28FVDD145 pKa = 5.58DD146 pKa = 4.63FWRR149 pKa = 11.84DD150 pKa = 3.31MGYY153 pKa = 10.08HH154 pKa = 5.52RR155 pKa = 11.84QARR158 pKa = 11.84TNPKK162 pKa = 9.63SLNFRR167 pKa = 11.84KK168 pKa = 9.29FHH170 pKa = 6.31LTTKK174 pKa = 10.19SGPNGTANALWTSLNDD190 pKa = 3.66LKK192 pKa = 11.24ALSEE196 pKa = 4.1QQLTDD201 pKa = 2.87IGIIGGEE208 pKa = 4.12TLRR211 pKa = 11.84SKK213 pKa = 10.51MSTLKK218 pKa = 10.74EE219 pKa = 4.12GLDD222 pKa = 3.66KK223 pKa = 11.37VPEE226 pKa = 4.05LSYY229 pKa = 11.39FMDD232 pKa = 3.98TSAGVYY238 pKa = 6.95TSRR241 pKa = 11.84MTSFPDD247 pKa = 3.06KK248 pKa = 9.45EE249 pKa = 4.23LKK251 pKa = 10.43VRR253 pKa = 11.84VIAVGDD259 pKa = 3.97YY260 pKa = 10.13FSQAALKK267 pKa = 9.53PLHH270 pKa = 7.32DD271 pKa = 3.79YY272 pKa = 11.0LFRR275 pKa = 11.84VLKK278 pKa = 10.23KK279 pKa = 10.22IPQDD283 pKa = 3.34CTFNQGGFWDD293 pKa = 4.38KK294 pKa = 11.11IKK296 pKa = 10.8DD297 pKa = 3.55QEE299 pKa = 4.54YY300 pKa = 9.82YY301 pKa = 11.37ASIDD305 pKa = 3.31LTAATDD311 pKa = 3.6SFPISTMSQVLLGSLPKK328 pKa = 10.33SYY330 pKa = 11.12VDD332 pKa = 2.73AWHH335 pKa = 7.01RR336 pKa = 11.84LMVGTPFIYY345 pKa = 10.12EE346 pKa = 4.12GQEE349 pKa = 3.34MNYY352 pKa = 10.83AVGNPMGFYY361 pKa = 10.68SSWASFAVAHH371 pKa = 6.5HH372 pKa = 5.41YY373 pKa = 9.52VVYY376 pKa = 9.96YY377 pKa = 9.81CCKK380 pKa = 10.41KK381 pKa = 10.84LGIPWKK387 pKa = 7.7TLKK390 pKa = 10.82YY391 pKa = 10.26CLLGDD396 pKa = 5.19DD397 pKa = 5.68IVMCDD402 pKa = 3.57PKK404 pKa = 11.13VAALYY409 pKa = 10.64KK410 pKa = 9.44EE411 pKa = 4.68TISGLGVEE419 pKa = 5.03FSEE422 pKa = 4.83EE423 pKa = 3.79KK424 pKa = 10.4TITSPHH430 pKa = 5.77LVEE433 pKa = 4.07FAKK436 pKa = 10.7RR437 pKa = 11.84LIYY440 pKa = 10.42KK441 pKa = 7.7GTEE444 pKa = 3.65MSPFPISALGEE455 pKa = 4.19LANKK459 pKa = 10.31YY460 pKa = 10.41YY461 pKa = 11.2LLTDD465 pKa = 4.19LFLEE469 pKa = 4.73LEE471 pKa = 4.37SKK473 pKa = 10.6GWVSLGGAPSAVEE486 pKa = 4.16SFHH489 pKa = 7.83KK490 pKa = 10.53IVFNWSTVRR499 pKa = 11.84ARR501 pKa = 11.84EE502 pKa = 4.04IEE504 pKa = 4.49VKK506 pKa = 10.73SSVCEE511 pKa = 3.75RR512 pKa = 11.84VMKK515 pKa = 9.47MARR518 pKa = 11.84GAEE521 pKa = 4.09DD522 pKa = 3.85AGTLLTEE529 pKa = 4.69CFSKK533 pKa = 10.77LGYY536 pKa = 9.88PFVLSSFVASNVLEE550 pKa = 4.09NMVVEE555 pKa = 4.94NFANSNPASHH565 pKa = 6.63NPEE568 pKa = 3.67SDD570 pKa = 3.39KK571 pKa = 11.04KK572 pKa = 11.03SKK574 pKa = 10.57SPCVGLGYY582 pKa = 10.25LAEE585 pKa = 4.65NLTCLLTGLDD595 pKa = 4.37EE596 pKa = 4.67EE597 pKa = 4.91SCSLGFDD604 pKa = 3.51LMMALPHH611 pKa = 6.3LNAYY615 pKa = 9.56GQMSEE620 pKa = 5.0LYY622 pKa = 11.09VNINKK627 pKa = 9.84KK628 pKa = 10.07ARR630 pKa = 11.84QISTTNGAEE639 pKa = 3.83WPLLFKK645 pKa = 9.71TMCIPWDD652 pKa = 3.53DD653 pKa = 4.76TIFTMSSSHH662 pKa = 7.14LIAKK666 pKa = 9.41GSSSVVKK673 pKa = 10.39DD674 pKa = 3.62LQNSAEE680 pKa = 4.43LLSFYY685 pKa = 10.49PPEE688 pKa = 3.96EE689 pKa = 4.18LLRR692 pKa = 11.84EE693 pKa = 4.31DD694 pKa = 4.05PNFLKK699 pKa = 10.93SLL701 pKa = 3.77
Molecular weight: 78.36 kDa
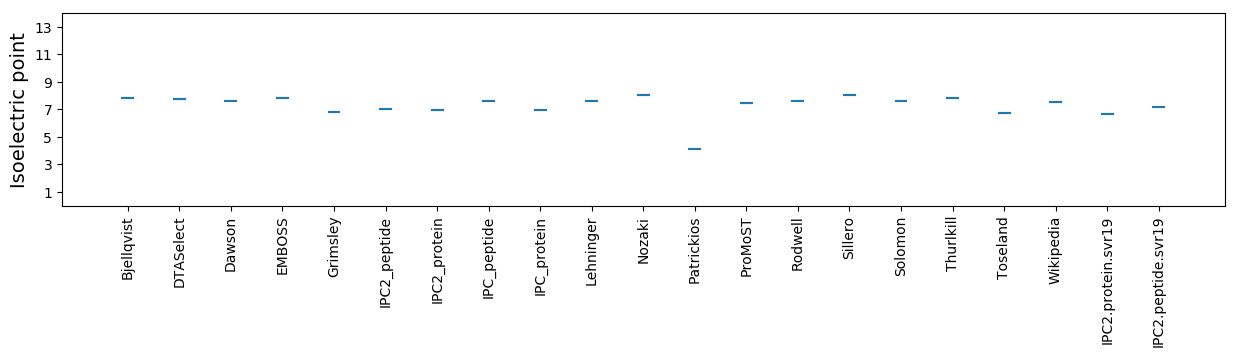
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KI82|A0A1L3KI82_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 25 OX=1922956 PE=4 SV=1
MM1 pKa = 7.64KK2 pKa = 7.96MTKK5 pKa = 10.37FNMFKK10 pKa = 9.54VTLKK14 pKa = 10.24VVTWMFQGFNLKK26 pKa = 9.7MSGRR30 pKa = 11.84MSDD33 pKa = 3.87YY34 pKa = 11.26LDD36 pKa = 5.73LILSLDD42 pKa = 3.35RR43 pKa = 11.84LLYY46 pKa = 10.6NRR48 pKa = 11.84GALEE52 pKa = 3.71TLKK55 pKa = 10.69YY56 pKa = 10.47IKK58 pKa = 9.74GTSTAYY64 pKa = 10.3LAYY67 pKa = 10.7LSGKK71 pKa = 7.64PMRR74 pKa = 11.84VKK76 pKa = 10.62GVSTTKK82 pKa = 10.76DD83 pKa = 3.82GIPVILGDD91 pKa = 4.19FVPKK95 pKa = 9.81IRR97 pKa = 11.84KK98 pKa = 9.3GPTPAMLQLINTILFCTSALKK119 pKa = 10.77LGSTPDD125 pKa = 4.1FSPMINPPKK134 pKa = 10.11SDD136 pKa = 4.77PINMGEE142 pKa = 4.28FVDD145 pKa = 5.58DD146 pKa = 4.63FWRR149 pKa = 11.84DD150 pKa = 3.31MGYY153 pKa = 10.08HH154 pKa = 5.52RR155 pKa = 11.84QARR158 pKa = 11.84TNPKK162 pKa = 9.63SLNFRR167 pKa = 11.84KK168 pKa = 9.29FHH170 pKa = 6.31LTTKK174 pKa = 10.19SGPNGTANALWTSLNDD190 pKa = 3.66LKK192 pKa = 11.24ALSEE196 pKa = 4.1QQLTDD201 pKa = 2.87IGIIGGEE208 pKa = 4.12TLRR211 pKa = 11.84SKK213 pKa = 10.51MSTLKK218 pKa = 10.74EE219 pKa = 4.12GLDD222 pKa = 3.66KK223 pKa = 11.37VPEE226 pKa = 4.05LSYY229 pKa = 11.39FMDD232 pKa = 3.98TSAGVYY238 pKa = 6.95TSRR241 pKa = 11.84MTSFPDD247 pKa = 3.06KK248 pKa = 9.45EE249 pKa = 4.23LKK251 pKa = 10.43VRR253 pKa = 11.84VIAVGDD259 pKa = 3.97YY260 pKa = 10.13FSQAALKK267 pKa = 9.53PLHH270 pKa = 7.32DD271 pKa = 3.79YY272 pKa = 11.0LFRR275 pKa = 11.84VLKK278 pKa = 10.23KK279 pKa = 10.22IPQDD283 pKa = 3.34CTFNQGGFWDD293 pKa = 4.38KK294 pKa = 11.11IKK296 pKa = 10.8DD297 pKa = 3.55QEE299 pKa = 4.54YY300 pKa = 9.82YY301 pKa = 11.37ASIDD305 pKa = 3.31LTAATDD311 pKa = 3.6SFPISTMSQVLLGSLPKK328 pKa = 10.33SYY330 pKa = 11.12VDD332 pKa = 2.73AWHH335 pKa = 7.01RR336 pKa = 11.84LMVGTPFIYY345 pKa = 10.12EE346 pKa = 4.12GQEE349 pKa = 3.34MNYY352 pKa = 10.83AVGNPMGFYY361 pKa = 10.68SSWASFAVAHH371 pKa = 6.5HH372 pKa = 5.41YY373 pKa = 9.52VVYY376 pKa = 9.96YY377 pKa = 9.81CCKK380 pKa = 10.41KK381 pKa = 10.84LGIPWKK387 pKa = 7.7TLKK390 pKa = 10.82YY391 pKa = 10.26CLLGDD396 pKa = 5.19DD397 pKa = 5.68IVMCDD402 pKa = 3.57PKK404 pKa = 11.13VAALYY409 pKa = 10.64KK410 pKa = 9.44EE411 pKa = 4.68TISGLGVEE419 pKa = 5.03FSEE422 pKa = 4.83EE423 pKa = 3.79KK424 pKa = 10.4TITSPHH430 pKa = 5.77LVEE433 pKa = 4.07FAKK436 pKa = 10.7RR437 pKa = 11.84LIYY440 pKa = 10.42KK441 pKa = 7.7GTEE444 pKa = 3.65MSPFPISALGEE455 pKa = 4.19LANKK459 pKa = 10.31YY460 pKa = 10.41YY461 pKa = 11.2LLTDD465 pKa = 4.19LFLEE469 pKa = 4.73LEE471 pKa = 4.37SKK473 pKa = 10.6GWVSLGGAPSAVEE486 pKa = 4.16SFHH489 pKa = 7.83KK490 pKa = 10.53IVFNWSTVRR499 pKa = 11.84ARR501 pKa = 11.84EE502 pKa = 4.04IEE504 pKa = 4.49VKK506 pKa = 10.73SSVCEE511 pKa = 3.75RR512 pKa = 11.84VMKK515 pKa = 9.47MARR518 pKa = 11.84GAEE521 pKa = 4.09DD522 pKa = 3.85AGTLLTEE529 pKa = 4.69CFSKK533 pKa = 10.77LGYY536 pKa = 9.88PFVLSSFVASNVLEE550 pKa = 4.09NMVVEE555 pKa = 4.94NFANSNPASHH565 pKa = 6.63NPEE568 pKa = 3.67SDD570 pKa = 3.39KK571 pKa = 11.04KK572 pKa = 11.03SKK574 pKa = 10.57SPCVGLGYY582 pKa = 10.25LAEE585 pKa = 4.65NLTCLLTGLDD595 pKa = 4.37EE596 pKa = 4.67EE597 pKa = 4.91SCSLGFDD604 pKa = 3.51LMMALPHH611 pKa = 6.3LNAYY615 pKa = 9.56GQMSEE620 pKa = 5.0LYY622 pKa = 11.09VNINKK627 pKa = 9.84KK628 pKa = 10.07ARR630 pKa = 11.84QISTTNGAEE639 pKa = 3.83WPLLFKK645 pKa = 9.71TMCIPWDD652 pKa = 3.53DD653 pKa = 4.76TIFTMSSSHH662 pKa = 7.14LIAKK666 pKa = 9.41GSSSVVKK673 pKa = 10.39DD674 pKa = 3.62LQNSAEE680 pKa = 4.43LLSFYY685 pKa = 10.49PPEE688 pKa = 3.96EE689 pKa = 4.18LLRR692 pKa = 11.84EE693 pKa = 4.31DD694 pKa = 4.05PNFLKK699 pKa = 10.93SLL701 pKa = 3.77
MM1 pKa = 7.64KK2 pKa = 7.96MTKK5 pKa = 10.37FNMFKK10 pKa = 9.54VTLKK14 pKa = 10.24VVTWMFQGFNLKK26 pKa = 9.7MSGRR30 pKa = 11.84MSDD33 pKa = 3.87YY34 pKa = 11.26LDD36 pKa = 5.73LILSLDD42 pKa = 3.35RR43 pKa = 11.84LLYY46 pKa = 10.6NRR48 pKa = 11.84GALEE52 pKa = 3.71TLKK55 pKa = 10.69YY56 pKa = 10.47IKK58 pKa = 9.74GTSTAYY64 pKa = 10.3LAYY67 pKa = 10.7LSGKK71 pKa = 7.64PMRR74 pKa = 11.84VKK76 pKa = 10.62GVSTTKK82 pKa = 10.76DD83 pKa = 3.82GIPVILGDD91 pKa = 4.19FVPKK95 pKa = 9.81IRR97 pKa = 11.84KK98 pKa = 9.3GPTPAMLQLINTILFCTSALKK119 pKa = 10.77LGSTPDD125 pKa = 4.1FSPMINPPKK134 pKa = 10.11SDD136 pKa = 4.77PINMGEE142 pKa = 4.28FVDD145 pKa = 5.58DD146 pKa = 4.63FWRR149 pKa = 11.84DD150 pKa = 3.31MGYY153 pKa = 10.08HH154 pKa = 5.52RR155 pKa = 11.84QARR158 pKa = 11.84TNPKK162 pKa = 9.63SLNFRR167 pKa = 11.84KK168 pKa = 9.29FHH170 pKa = 6.31LTTKK174 pKa = 10.19SGPNGTANALWTSLNDD190 pKa = 3.66LKK192 pKa = 11.24ALSEE196 pKa = 4.1QQLTDD201 pKa = 2.87IGIIGGEE208 pKa = 4.12TLRR211 pKa = 11.84SKK213 pKa = 10.51MSTLKK218 pKa = 10.74EE219 pKa = 4.12GLDD222 pKa = 3.66KK223 pKa = 11.37VPEE226 pKa = 4.05LSYY229 pKa = 11.39FMDD232 pKa = 3.98TSAGVYY238 pKa = 6.95TSRR241 pKa = 11.84MTSFPDD247 pKa = 3.06KK248 pKa = 9.45EE249 pKa = 4.23LKK251 pKa = 10.43VRR253 pKa = 11.84VIAVGDD259 pKa = 3.97YY260 pKa = 10.13FSQAALKK267 pKa = 9.53PLHH270 pKa = 7.32DD271 pKa = 3.79YY272 pKa = 11.0LFRR275 pKa = 11.84VLKK278 pKa = 10.23KK279 pKa = 10.22IPQDD283 pKa = 3.34CTFNQGGFWDD293 pKa = 4.38KK294 pKa = 11.11IKK296 pKa = 10.8DD297 pKa = 3.55QEE299 pKa = 4.54YY300 pKa = 9.82YY301 pKa = 11.37ASIDD305 pKa = 3.31LTAATDD311 pKa = 3.6SFPISTMSQVLLGSLPKK328 pKa = 10.33SYY330 pKa = 11.12VDD332 pKa = 2.73AWHH335 pKa = 7.01RR336 pKa = 11.84LMVGTPFIYY345 pKa = 10.12EE346 pKa = 4.12GQEE349 pKa = 3.34MNYY352 pKa = 10.83AVGNPMGFYY361 pKa = 10.68SSWASFAVAHH371 pKa = 6.5HH372 pKa = 5.41YY373 pKa = 9.52VVYY376 pKa = 9.96YY377 pKa = 9.81CCKK380 pKa = 10.41KK381 pKa = 10.84LGIPWKK387 pKa = 7.7TLKK390 pKa = 10.82YY391 pKa = 10.26CLLGDD396 pKa = 5.19DD397 pKa = 5.68IVMCDD402 pKa = 3.57PKK404 pKa = 11.13VAALYY409 pKa = 10.64KK410 pKa = 9.44EE411 pKa = 4.68TISGLGVEE419 pKa = 5.03FSEE422 pKa = 4.83EE423 pKa = 3.79KK424 pKa = 10.4TITSPHH430 pKa = 5.77LVEE433 pKa = 4.07FAKK436 pKa = 10.7RR437 pKa = 11.84LIYY440 pKa = 10.42KK441 pKa = 7.7GTEE444 pKa = 3.65MSPFPISALGEE455 pKa = 4.19LANKK459 pKa = 10.31YY460 pKa = 10.41YY461 pKa = 11.2LLTDD465 pKa = 4.19LFLEE469 pKa = 4.73LEE471 pKa = 4.37SKK473 pKa = 10.6GWVSLGGAPSAVEE486 pKa = 4.16SFHH489 pKa = 7.83KK490 pKa = 10.53IVFNWSTVRR499 pKa = 11.84ARR501 pKa = 11.84EE502 pKa = 4.04IEE504 pKa = 4.49VKK506 pKa = 10.73SSVCEE511 pKa = 3.75RR512 pKa = 11.84VMKK515 pKa = 9.47MARR518 pKa = 11.84GAEE521 pKa = 4.09DD522 pKa = 3.85AGTLLTEE529 pKa = 4.69CFSKK533 pKa = 10.77LGYY536 pKa = 9.88PFVLSSFVASNVLEE550 pKa = 4.09NMVVEE555 pKa = 4.94NFANSNPASHH565 pKa = 6.63NPEE568 pKa = 3.67SDD570 pKa = 3.39KK571 pKa = 11.04KK572 pKa = 11.03SKK574 pKa = 10.57SPCVGLGYY582 pKa = 10.25LAEE585 pKa = 4.65NLTCLLTGLDD595 pKa = 4.37EE596 pKa = 4.67EE597 pKa = 4.91SCSLGFDD604 pKa = 3.51LMMALPHH611 pKa = 6.3LNAYY615 pKa = 9.56GQMSEE620 pKa = 5.0LYY622 pKa = 11.09VNINKK627 pKa = 9.84KK628 pKa = 10.07ARR630 pKa = 11.84QISTTNGAEE639 pKa = 3.83WPLLFKK645 pKa = 9.71TMCIPWDD652 pKa = 3.53DD653 pKa = 4.76TIFTMSSSHH662 pKa = 7.14LIAKK666 pKa = 9.41GSSSVVKK673 pKa = 10.39DD674 pKa = 3.62LQNSAEE680 pKa = 4.43LLSFYY685 pKa = 10.49PPEE688 pKa = 3.96EE689 pKa = 4.18LLRR692 pKa = 11.84EE693 pKa = 4.31DD694 pKa = 4.05PNFLKK699 pKa = 10.93SLL701 pKa = 3.77
Molecular weight: 78.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
701 |
701 |
701 |
701.0 |
78.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.991 ± 0.0 | 1.712 ± 0.0 |
4.993 ± 0.0 | 5.278 ± 0.0 |
5.136 ± 0.0 | 6.562 ± 0.0 |
1.569 ± 0.0 | 4.422 ± 0.0 |
7.561 ± 0.0 | 11.555 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.994 ± 0.0 | 4.137 ± 0.0 |
5.136 ± 0.0 | 1.997 ± 0.0 |
2.996 ± 0.0 | 9.13 ± 0.0 |
6.277 ± 0.0 | 5.849 ± 0.0 |
1.569 ± 0.0 | 4.137 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
