
Anabaena sp. 90
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Nostocales; Nostocaceae; Anabaena; unclassified Anabaena
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
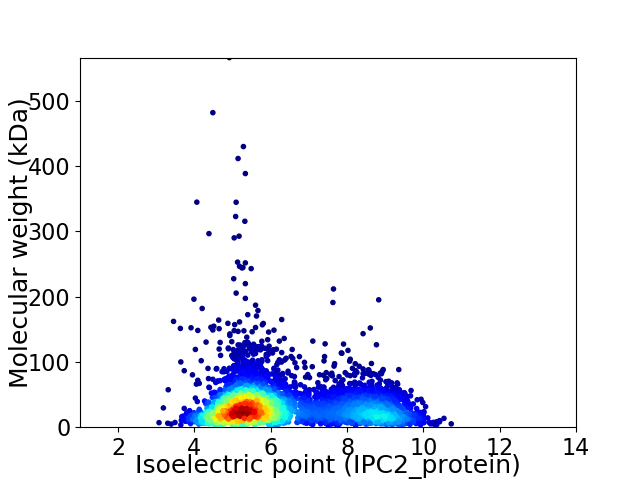
Virtual 2D-PAGE plot for 4476 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K7WVR6|K7WVR6_9NOST Universal stress protein UspA OS=Anabaena sp. 90 OX=46234 GN=uspA PE=3 SV=1
MM1 pKa = 7.22SQQNLVFTNPNEE13 pKa = 3.8ITIPNGGAATTYY25 pKa = 9.43PSSINVSGLNGTITSLKK42 pKa = 8.6VTLSNISHH50 pKa = 7.46TYY52 pKa = 10.27PDD54 pKa = 4.79DD55 pKa = 5.24LDD57 pKa = 4.09ILLIGPTGANILLMSDD73 pKa = 5.1AGWSWDD79 pKa = 3.77LNNVTLTFDD88 pKa = 3.9PNASDD93 pKa = 4.28FLPDD97 pKa = 3.09QGQISSGSYY106 pKa = 9.51RR107 pKa = 11.84ATDD110 pKa = 3.5YY111 pKa = 9.05EE112 pKa = 4.06TDD114 pKa = 3.47DD115 pKa = 4.17RR116 pKa = 11.84FNNSPAPSGDD126 pKa = 3.36YY127 pKa = 11.3GIDD130 pKa = 3.12FSVFNNTIPNGTWNLYY146 pKa = 9.86VVDD149 pKa = 4.4DD150 pKa = 4.12TGVDD154 pKa = 2.96IGNIAGGWSIAIDD167 pKa = 3.73TTDD170 pKa = 3.18VTTKK174 pKa = 10.52PIVTLATSPKK184 pKa = 9.89KK185 pKa = 10.22VAEE188 pKa = 4.11HH189 pKa = 6.13SGGNILYY196 pKa = 8.45TFTRR200 pKa = 11.84TGNITSALTVNYY212 pKa = 7.95TVSGSAIFNTDD223 pKa = 2.99YY224 pKa = 10.97SQTGATSHH232 pKa = 6.31TATTGTVSFAAGADD246 pKa = 3.75TVTLTISPLEE256 pKa = 4.32DD257 pKa = 3.91NIQEE261 pKa = 3.97NDD263 pKa = 3.04EE264 pKa = 4.49TIILTLATATDD275 pKa = 4.38YY276 pKa = 11.1IIGTTTEE283 pKa = 3.94LTGTITNGNDD293 pKa = 2.78QFEE296 pKa = 4.67ATGSNLVSNKK306 pKa = 9.69IYY308 pKa = 10.67DD309 pKa = 3.9GGGGTDD315 pKa = 3.46TLIVNYY321 pKa = 10.44SSATFSGLGTDD332 pKa = 4.31GLGVSNGYY340 pKa = 8.85WSNSAFHH347 pKa = 7.83DD348 pKa = 3.68YY349 pKa = 10.61RR350 pKa = 11.84GYY352 pKa = 11.21YY353 pKa = 8.81YY354 pKa = 10.57YY355 pKa = 11.09GWRR358 pKa = 11.84ANSPLLSFSNIEE370 pKa = 4.0TFEE373 pKa = 4.16ITGTAYY379 pKa = 10.37ADD381 pKa = 3.66YY382 pKa = 10.96LRR384 pKa = 11.84GYY386 pKa = 10.34SGDD389 pKa = 3.78DD390 pKa = 3.26KK391 pKa = 11.65LIGGEE396 pKa = 4.32GNDD399 pKa = 5.23DD400 pKa = 3.39IAGGDD405 pKa = 3.84GNDD408 pKa = 5.02LIRR411 pKa = 11.84GGDD414 pKa = 3.69GNDD417 pKa = 3.33TLKK420 pKa = 11.45GEE422 pKa = 4.43GGNDD426 pKa = 3.49EE427 pKa = 4.85IEE429 pKa = 4.71GGDD432 pKa = 3.99GNDD435 pKa = 4.2TIDD438 pKa = 3.72GGTGRR443 pKa = 11.84HH444 pKa = 5.61VIRR447 pKa = 11.84GGTGNDD453 pKa = 2.98TVAGAKK459 pKa = 10.36AGDD462 pKa = 4.27SIDD465 pKa = 4.17GEE467 pKa = 4.62GGNDD471 pKa = 3.42TLQSLDD477 pKa = 3.83LSSLGVNLVLDD488 pKa = 4.29LRR490 pKa = 11.84ATEE493 pKa = 4.25QVSGDD498 pKa = 3.16NNTRR502 pKa = 11.84INNFEE507 pKa = 4.63NISSVTLGSGHH518 pKa = 7.23DD519 pKa = 3.6TTIATSNLLVSNGSINAGGGTDD541 pKa = 3.8RR542 pKa = 11.84LIVDD546 pKa = 3.73YY547 pKa = 11.33SSATFSGLGTDD558 pKa = 4.31GLGVSNGYY566 pKa = 8.85WSNSAFHH573 pKa = 7.83DD574 pKa = 3.68YY575 pKa = 10.61RR576 pKa = 11.84GYY578 pKa = 11.21YY579 pKa = 8.81YY580 pKa = 10.57YY581 pKa = 11.09GWRR584 pKa = 11.84ANSPLLSFSNIEE596 pKa = 4.0TFEE599 pKa = 4.16ITGTAYY605 pKa = 10.37ADD607 pKa = 3.66YY608 pKa = 10.96LRR610 pKa = 11.84GYY612 pKa = 10.34SGDD615 pKa = 3.78DD616 pKa = 3.26KK617 pKa = 11.65LIGGEE622 pKa = 4.32GNDD625 pKa = 5.23DD626 pKa = 3.39IAGGDD631 pKa = 3.84GNDD634 pKa = 5.02LIRR637 pKa = 11.84GGDD640 pKa = 3.69GNDD643 pKa = 3.33TLKK646 pKa = 11.45GEE648 pKa = 4.43GGNDD652 pKa = 3.49EE653 pKa = 4.85IEE655 pKa = 4.71GGDD658 pKa = 3.99GNDD661 pKa = 4.2TIDD664 pKa = 3.78GGTGRR669 pKa = 11.84HH670 pKa = 6.9LIRR673 pKa = 11.84GGKK676 pKa = 10.1GDD678 pKa = 3.92DD679 pKa = 3.45TVVSAKK685 pKa = 10.69AGDD688 pKa = 4.28SIDD691 pKa = 3.74GGEE694 pKa = 4.76GNDD697 pKa = 3.73TLQSLDD703 pKa = 4.14LSSLDD708 pKa = 3.16VDD710 pKa = 4.29LVLNLRR716 pKa = 11.84VTEE719 pKa = 4.01QLSGDD724 pKa = 3.37NNTRR728 pKa = 11.84INNFEE733 pKa = 4.55NISTVTLGSGYY744 pKa = 8.83DD745 pKa = 3.36TTIATGNLLVSNGSFNGGGGTDD767 pKa = 3.65TLIVDD772 pKa = 3.87YY773 pKa = 11.3SSATFSGLGTDD784 pKa = 4.31GLGVSNGYY792 pKa = 8.85WSNSAFHH799 pKa = 7.83DD800 pKa = 3.68YY801 pKa = 10.61RR802 pKa = 11.84GYY804 pKa = 11.21YY805 pKa = 8.81YY806 pKa = 10.57YY807 pKa = 11.09GWRR810 pKa = 11.84ANSPLLSFSNIEE822 pKa = 4.0TFEE825 pKa = 4.16ITGTAYY831 pKa = 10.37ADD833 pKa = 3.66YY834 pKa = 10.96LRR836 pKa = 11.84GYY838 pKa = 10.34SGDD841 pKa = 3.78DD842 pKa = 3.26KK843 pKa = 11.65LIGGEE848 pKa = 4.27GNDD851 pKa = 4.26NIAGGDD857 pKa = 3.65GSDD860 pKa = 3.89TIIPVNPNVAKK871 pKa = 10.42PGLGTVDD878 pKa = 3.48TVTGGTGEE886 pKa = 4.15DD887 pKa = 3.61TFILGDD893 pKa = 3.97ATWIGYY899 pKa = 9.95DD900 pKa = 4.32DD901 pKa = 4.49YY902 pKa = 12.02NSTTAGTSDD911 pKa = 3.41YY912 pKa = 11.26LIIKK916 pKa = 9.25DD917 pKa = 4.26FNPLEE922 pKa = 5.49DD923 pKa = 4.57IIQLLGSSSDD933 pKa = 3.52YY934 pKa = 8.07TTTVSGNNINLYY946 pKa = 9.87INKK949 pKa = 8.93PDD951 pKa = 3.84SEE953 pKa = 4.53PDD955 pKa = 3.07EE956 pKa = 5.1LIAVLEE962 pKa = 4.17NTQGLINSLQFNLTGSYY979 pKa = 10.26FKK981 pKa = 11.04YY982 pKa = 9.66IASNAVLNITSTSTSRR998 pKa = 11.84IQTEE1002 pKa = 3.99GDD1004 pKa = 3.28SDD1006 pKa = 3.61NKK1008 pKa = 10.96SFVFTVTRR1016 pKa = 11.84TGDD1019 pKa = 3.12ASGTSTVDD1027 pKa = 2.88WFITPSGDD1035 pKa = 3.36NPVNSTDD1042 pKa = 3.89FFGGAIPPGEE1052 pKa = 4.41SLTFLPGEE1060 pKa = 3.98TSKK1063 pKa = 10.86EE1064 pKa = 3.27IRR1066 pKa = 11.84VRR1068 pKa = 11.84VRR1070 pKa = 11.84GDD1072 pKa = 3.14SAIEE1076 pKa = 4.05SNEE1079 pKa = 3.7TFTVYY1084 pKa = 10.7LSTPSRR1090 pKa = 11.84STIGTRR1096 pKa = 11.84SATGIILNDD1105 pKa = 4.64DD1106 pKa = 4.13GSQPTLPVIGTSGNDD1121 pKa = 3.58TLSGTINADD1130 pKa = 3.56CLIGLAGNDD1139 pKa = 3.3TYY1141 pKa = 11.32TVNNAGDD1148 pKa = 3.78VVVEE1152 pKa = 4.32NPSEE1156 pKa = 4.0GTDD1159 pKa = 2.94RR1160 pKa = 11.84VNASISYY1167 pKa = 8.49TLADD1171 pKa = 3.38NVEE1174 pKa = 4.07NLTLTGSANLNGTGNDD1190 pKa = 3.96LNNSLTGNTGNNILTGNAGNDD1211 pKa = 3.92TLNGAAGIDD1220 pKa = 3.75TLIGGLGDD1228 pKa = 4.75DD1229 pKa = 4.26IYY1231 pKa = 11.77VVDD1234 pKa = 4.46STTDD1238 pKa = 3.39TITEE1242 pKa = 4.03NSGEE1246 pKa = 4.17GTDD1249 pKa = 4.41IIQSSVTFDD1258 pKa = 3.04LTVFPNIEE1266 pKa = 4.16NLTLTGTAVINGTGNAGDD1284 pKa = 3.82NSLTGNSGNNTLTGNAGNDD1303 pKa = 3.77LLNGGTGNDD1312 pKa = 4.3LLNGGAGADD1321 pKa = 3.8TLTGGIGSDD1330 pKa = 3.13TLVFQFGQSPVSGADD1345 pKa = 3.31RR1346 pKa = 11.84ITDD1349 pKa = 3.92FAIGTDD1355 pKa = 4.05KK1356 pKa = 10.9IDD1358 pKa = 3.9LLTSLGVAMNAPTAFTRR1375 pKa = 11.84AANSTDD1381 pKa = 3.17TTLTNVVNSVFTDD1394 pKa = 3.36ANGALTDD1401 pKa = 3.81NQALGVSSAALVSVTTSGIAGTYY1424 pKa = 10.11LVINDD1429 pKa = 4.34GVAGFQSSNDD1439 pKa = 3.44LLVNITGSSGTLPALGTIAVSSFFII1464 pKa = 4.76
MM1 pKa = 7.22SQQNLVFTNPNEE13 pKa = 3.8ITIPNGGAATTYY25 pKa = 9.43PSSINVSGLNGTITSLKK42 pKa = 8.6VTLSNISHH50 pKa = 7.46TYY52 pKa = 10.27PDD54 pKa = 4.79DD55 pKa = 5.24LDD57 pKa = 4.09ILLIGPTGANILLMSDD73 pKa = 5.1AGWSWDD79 pKa = 3.77LNNVTLTFDD88 pKa = 3.9PNASDD93 pKa = 4.28FLPDD97 pKa = 3.09QGQISSGSYY106 pKa = 9.51RR107 pKa = 11.84ATDD110 pKa = 3.5YY111 pKa = 9.05EE112 pKa = 4.06TDD114 pKa = 3.47DD115 pKa = 4.17RR116 pKa = 11.84FNNSPAPSGDD126 pKa = 3.36YY127 pKa = 11.3GIDD130 pKa = 3.12FSVFNNTIPNGTWNLYY146 pKa = 9.86VVDD149 pKa = 4.4DD150 pKa = 4.12TGVDD154 pKa = 2.96IGNIAGGWSIAIDD167 pKa = 3.73TTDD170 pKa = 3.18VTTKK174 pKa = 10.52PIVTLATSPKK184 pKa = 9.89KK185 pKa = 10.22VAEE188 pKa = 4.11HH189 pKa = 6.13SGGNILYY196 pKa = 8.45TFTRR200 pKa = 11.84TGNITSALTVNYY212 pKa = 7.95TVSGSAIFNTDD223 pKa = 2.99YY224 pKa = 10.97SQTGATSHH232 pKa = 6.31TATTGTVSFAAGADD246 pKa = 3.75TVTLTISPLEE256 pKa = 4.32DD257 pKa = 3.91NIQEE261 pKa = 3.97NDD263 pKa = 3.04EE264 pKa = 4.49TIILTLATATDD275 pKa = 4.38YY276 pKa = 11.1IIGTTTEE283 pKa = 3.94LTGTITNGNDD293 pKa = 2.78QFEE296 pKa = 4.67ATGSNLVSNKK306 pKa = 9.69IYY308 pKa = 10.67DD309 pKa = 3.9GGGGTDD315 pKa = 3.46TLIVNYY321 pKa = 10.44SSATFSGLGTDD332 pKa = 4.31GLGVSNGYY340 pKa = 8.85WSNSAFHH347 pKa = 7.83DD348 pKa = 3.68YY349 pKa = 10.61RR350 pKa = 11.84GYY352 pKa = 11.21YY353 pKa = 8.81YY354 pKa = 10.57YY355 pKa = 11.09GWRR358 pKa = 11.84ANSPLLSFSNIEE370 pKa = 4.0TFEE373 pKa = 4.16ITGTAYY379 pKa = 10.37ADD381 pKa = 3.66YY382 pKa = 10.96LRR384 pKa = 11.84GYY386 pKa = 10.34SGDD389 pKa = 3.78DD390 pKa = 3.26KK391 pKa = 11.65LIGGEE396 pKa = 4.32GNDD399 pKa = 5.23DD400 pKa = 3.39IAGGDD405 pKa = 3.84GNDD408 pKa = 5.02LIRR411 pKa = 11.84GGDD414 pKa = 3.69GNDD417 pKa = 3.33TLKK420 pKa = 11.45GEE422 pKa = 4.43GGNDD426 pKa = 3.49EE427 pKa = 4.85IEE429 pKa = 4.71GGDD432 pKa = 3.99GNDD435 pKa = 4.2TIDD438 pKa = 3.72GGTGRR443 pKa = 11.84HH444 pKa = 5.61VIRR447 pKa = 11.84GGTGNDD453 pKa = 2.98TVAGAKK459 pKa = 10.36AGDD462 pKa = 4.27SIDD465 pKa = 4.17GEE467 pKa = 4.62GGNDD471 pKa = 3.42TLQSLDD477 pKa = 3.83LSSLGVNLVLDD488 pKa = 4.29LRR490 pKa = 11.84ATEE493 pKa = 4.25QVSGDD498 pKa = 3.16NNTRR502 pKa = 11.84INNFEE507 pKa = 4.63NISSVTLGSGHH518 pKa = 7.23DD519 pKa = 3.6TTIATSNLLVSNGSINAGGGTDD541 pKa = 3.8RR542 pKa = 11.84LIVDD546 pKa = 3.73YY547 pKa = 11.33SSATFSGLGTDD558 pKa = 4.31GLGVSNGYY566 pKa = 8.85WSNSAFHH573 pKa = 7.83DD574 pKa = 3.68YY575 pKa = 10.61RR576 pKa = 11.84GYY578 pKa = 11.21YY579 pKa = 8.81YY580 pKa = 10.57YY581 pKa = 11.09GWRR584 pKa = 11.84ANSPLLSFSNIEE596 pKa = 4.0TFEE599 pKa = 4.16ITGTAYY605 pKa = 10.37ADD607 pKa = 3.66YY608 pKa = 10.96LRR610 pKa = 11.84GYY612 pKa = 10.34SGDD615 pKa = 3.78DD616 pKa = 3.26KK617 pKa = 11.65LIGGEE622 pKa = 4.32GNDD625 pKa = 5.23DD626 pKa = 3.39IAGGDD631 pKa = 3.84GNDD634 pKa = 5.02LIRR637 pKa = 11.84GGDD640 pKa = 3.69GNDD643 pKa = 3.33TLKK646 pKa = 11.45GEE648 pKa = 4.43GGNDD652 pKa = 3.49EE653 pKa = 4.85IEE655 pKa = 4.71GGDD658 pKa = 3.99GNDD661 pKa = 4.2TIDD664 pKa = 3.78GGTGRR669 pKa = 11.84HH670 pKa = 6.9LIRR673 pKa = 11.84GGKK676 pKa = 10.1GDD678 pKa = 3.92DD679 pKa = 3.45TVVSAKK685 pKa = 10.69AGDD688 pKa = 4.28SIDD691 pKa = 3.74GGEE694 pKa = 4.76GNDD697 pKa = 3.73TLQSLDD703 pKa = 4.14LSSLDD708 pKa = 3.16VDD710 pKa = 4.29LVLNLRR716 pKa = 11.84VTEE719 pKa = 4.01QLSGDD724 pKa = 3.37NNTRR728 pKa = 11.84INNFEE733 pKa = 4.55NISTVTLGSGYY744 pKa = 8.83DD745 pKa = 3.36TTIATGNLLVSNGSFNGGGGTDD767 pKa = 3.65TLIVDD772 pKa = 3.87YY773 pKa = 11.3SSATFSGLGTDD784 pKa = 4.31GLGVSNGYY792 pKa = 8.85WSNSAFHH799 pKa = 7.83DD800 pKa = 3.68YY801 pKa = 10.61RR802 pKa = 11.84GYY804 pKa = 11.21YY805 pKa = 8.81YY806 pKa = 10.57YY807 pKa = 11.09GWRR810 pKa = 11.84ANSPLLSFSNIEE822 pKa = 4.0TFEE825 pKa = 4.16ITGTAYY831 pKa = 10.37ADD833 pKa = 3.66YY834 pKa = 10.96LRR836 pKa = 11.84GYY838 pKa = 10.34SGDD841 pKa = 3.78DD842 pKa = 3.26KK843 pKa = 11.65LIGGEE848 pKa = 4.27GNDD851 pKa = 4.26NIAGGDD857 pKa = 3.65GSDD860 pKa = 3.89TIIPVNPNVAKK871 pKa = 10.42PGLGTVDD878 pKa = 3.48TVTGGTGEE886 pKa = 4.15DD887 pKa = 3.61TFILGDD893 pKa = 3.97ATWIGYY899 pKa = 9.95DD900 pKa = 4.32DD901 pKa = 4.49YY902 pKa = 12.02NSTTAGTSDD911 pKa = 3.41YY912 pKa = 11.26LIIKK916 pKa = 9.25DD917 pKa = 4.26FNPLEE922 pKa = 5.49DD923 pKa = 4.57IIQLLGSSSDD933 pKa = 3.52YY934 pKa = 8.07TTTVSGNNINLYY946 pKa = 9.87INKK949 pKa = 8.93PDD951 pKa = 3.84SEE953 pKa = 4.53PDD955 pKa = 3.07EE956 pKa = 5.1LIAVLEE962 pKa = 4.17NTQGLINSLQFNLTGSYY979 pKa = 10.26FKK981 pKa = 11.04YY982 pKa = 9.66IASNAVLNITSTSTSRR998 pKa = 11.84IQTEE1002 pKa = 3.99GDD1004 pKa = 3.28SDD1006 pKa = 3.61NKK1008 pKa = 10.96SFVFTVTRR1016 pKa = 11.84TGDD1019 pKa = 3.12ASGTSTVDD1027 pKa = 2.88WFITPSGDD1035 pKa = 3.36NPVNSTDD1042 pKa = 3.89FFGGAIPPGEE1052 pKa = 4.41SLTFLPGEE1060 pKa = 3.98TSKK1063 pKa = 10.86EE1064 pKa = 3.27IRR1066 pKa = 11.84VRR1068 pKa = 11.84VRR1070 pKa = 11.84GDD1072 pKa = 3.14SAIEE1076 pKa = 4.05SNEE1079 pKa = 3.7TFTVYY1084 pKa = 10.7LSTPSRR1090 pKa = 11.84STIGTRR1096 pKa = 11.84SATGIILNDD1105 pKa = 4.64DD1106 pKa = 4.13GSQPTLPVIGTSGNDD1121 pKa = 3.58TLSGTINADD1130 pKa = 3.56CLIGLAGNDD1139 pKa = 3.3TYY1141 pKa = 11.32TVNNAGDD1148 pKa = 3.78VVVEE1152 pKa = 4.32NPSEE1156 pKa = 4.0GTDD1159 pKa = 2.94RR1160 pKa = 11.84VNASISYY1167 pKa = 8.49TLADD1171 pKa = 3.38NVEE1174 pKa = 4.07NLTLTGSANLNGTGNDD1190 pKa = 3.96LNNSLTGNTGNNILTGNAGNDD1211 pKa = 3.92TLNGAAGIDD1220 pKa = 3.75TLIGGLGDD1228 pKa = 4.75DD1229 pKa = 4.26IYY1231 pKa = 11.77VVDD1234 pKa = 4.46STTDD1238 pKa = 3.39TITEE1242 pKa = 4.03NSGEE1246 pKa = 4.17GTDD1249 pKa = 4.41IIQSSVTFDD1258 pKa = 3.04LTVFPNIEE1266 pKa = 4.16NLTLTGTAVINGTGNAGDD1284 pKa = 3.82NSLTGNSGNNTLTGNAGNDD1303 pKa = 3.77LLNGGTGNDD1312 pKa = 4.3LLNGGAGADD1321 pKa = 3.8TLTGGIGSDD1330 pKa = 3.13TLVFQFGQSPVSGADD1345 pKa = 3.31RR1346 pKa = 11.84ITDD1349 pKa = 3.92FAIGTDD1355 pKa = 4.05KK1356 pKa = 10.9IDD1358 pKa = 3.9LLTSLGVAMNAPTAFTRR1375 pKa = 11.84AANSTDD1381 pKa = 3.17TTLTNVVNSVFTDD1394 pKa = 3.36ANGALTDD1401 pKa = 3.81NQALGVSSAALVSVTTSGIAGTYY1424 pKa = 10.11LVINDD1429 pKa = 4.34GVAGFQSSNDD1439 pKa = 3.44LLVNITGSSGTLPALGTIAVSSFFII1464 pKa = 4.76
Molecular weight: 151.42 kDa
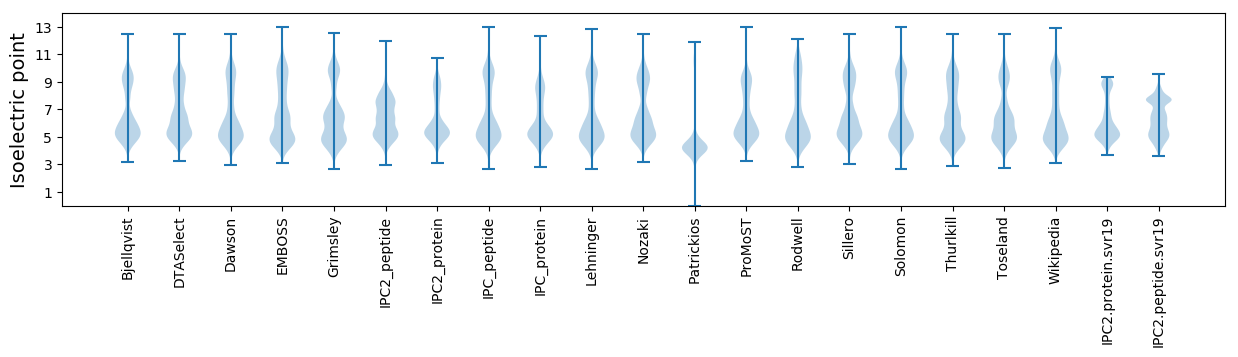
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7W0S5|K7W0S5_9NOST Non-specific serine/threonine protein kinase OS=Anabaena sp. 90 OX=46234 GN=ANA_C13407 PE=4 SV=1
MM1 pKa = 6.44QQRR4 pKa = 11.84FAIASLQLPKK14 pKa = 10.17SDD16 pKa = 5.19RR17 pKa = 11.84IPSTSQTANCLQQRR31 pKa = 11.84FAIAPPQPSKK41 pKa = 10.46QRR43 pKa = 11.84RR44 pKa = 11.84TPSTSQTANCLQQRR58 pKa = 11.84FAIAPLNPPNSEE70 pKa = 4.1LLAAALRR77 pKa = 11.84YY78 pKa = 7.67RR79 pKa = 11.84TPQPPKK85 pKa = 10.37QRR87 pKa = 11.84TACSSASLSHH97 pKa = 7.03PLNLPNSEE105 pKa = 4.46LLAAALRR112 pKa = 11.84YY113 pKa = 7.53RR114 pKa = 11.84TSSNFPNILFLTNHH128 pKa = 7.15RR129 pKa = 11.84DD130 pKa = 3.54TEE132 pKa = 4.25DD133 pKa = 3.52TEE135 pKa = 4.63KK136 pKa = 10.84EE137 pKa = 4.05RR138 pKa = 11.84SHH140 pKa = 7.33SLL142 pKa = 3.18
MM1 pKa = 6.44QQRR4 pKa = 11.84FAIASLQLPKK14 pKa = 10.17SDD16 pKa = 5.19RR17 pKa = 11.84IPSTSQTANCLQQRR31 pKa = 11.84FAIAPPQPSKK41 pKa = 10.46QRR43 pKa = 11.84RR44 pKa = 11.84TPSTSQTANCLQQRR58 pKa = 11.84FAIAPLNPPNSEE70 pKa = 4.1LLAAALRR77 pKa = 11.84YY78 pKa = 7.67RR79 pKa = 11.84TPQPPKK85 pKa = 10.37QRR87 pKa = 11.84TACSSASLSHH97 pKa = 7.03PLNLPNSEE105 pKa = 4.46LLAAALRR112 pKa = 11.84YY113 pKa = 7.53RR114 pKa = 11.84TSSNFPNILFLTNHH128 pKa = 7.15RR129 pKa = 11.84DD130 pKa = 3.54TEE132 pKa = 4.25DD133 pKa = 3.52TEE135 pKa = 4.63KK136 pKa = 10.84EE137 pKa = 4.05RR138 pKa = 11.84SHH140 pKa = 7.33SLL142 pKa = 3.18
Molecular weight: 15.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1402120 |
28 |
5062 |
313.3 |
35.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.349 ± 0.036 | 0.995 ± 0.013 |
4.938 ± 0.025 | 6.363 ± 0.034 |
4.036 ± 0.024 | 6.446 ± 0.036 |
1.843 ± 0.017 | 7.498 ± 0.03 |
5.494 ± 0.039 | 10.836 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.842 ± 0.019 | 5.012 ± 0.036 |
4.53 ± 0.024 | 5.305 ± 0.037 |
4.569 ± 0.029 | 6.35 ± 0.028 |
5.741 ± 0.037 | 6.27 ± 0.03 |
1.403 ± 0.016 | 3.182 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
