
Canis familiaris papillomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Taupapillomavirus; Taupapillomavirus 1
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
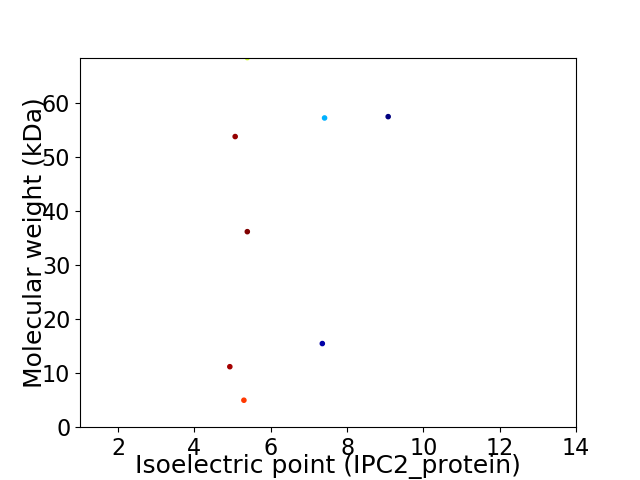
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
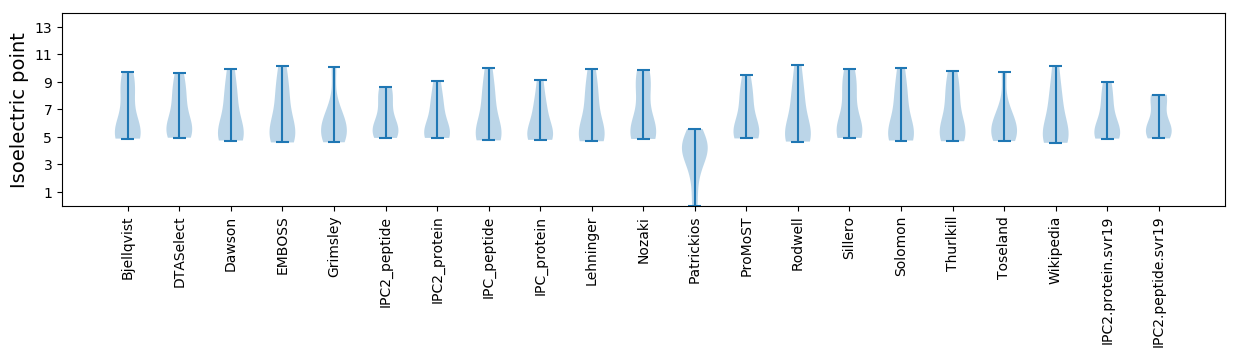
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q647I1|Q647I1_9PAPI Protein E6 OS=Canis familiaris papillomavirus 2 OX=292792 GN=E6 PE=3 SV=1
MM1 pKa = 7.73RR2 pKa = 11.84GSSPIIRR9 pKa = 11.84DD10 pKa = 2.73IDD12 pKa = 3.83LEE14 pKa = 4.26LEE16 pKa = 4.24EE17 pKa = 5.38LVLPANLLSGEE28 pKa = 4.16TLEE31 pKa = 4.39TEE33 pKa = 4.15EE34 pKa = 5.5EE35 pKa = 4.08EE36 pKa = 4.42LQRR39 pKa = 11.84EE40 pKa = 4.23PGRR43 pKa = 11.84YY44 pKa = 8.22RR45 pKa = 11.84VVTLCNICHH54 pKa = 6.02SSLRR58 pKa = 11.84LFVEE62 pKa = 4.47VADD65 pKa = 4.31EE66 pKa = 4.03SLIRR70 pKa = 11.84LFQQLLLDD78 pKa = 4.0GLGIICATCHH88 pKa = 6.56KK89 pKa = 10.12EE90 pKa = 3.8HH91 pKa = 6.88FSDD94 pKa = 3.74GRR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 3.42
MM1 pKa = 7.73RR2 pKa = 11.84GSSPIIRR9 pKa = 11.84DD10 pKa = 2.73IDD12 pKa = 3.83LEE14 pKa = 4.26LEE16 pKa = 4.24EE17 pKa = 5.38LVLPANLLSGEE28 pKa = 4.16TLEE31 pKa = 4.39TEE33 pKa = 4.15EE34 pKa = 5.5EE35 pKa = 4.08EE36 pKa = 4.42LQRR39 pKa = 11.84EE40 pKa = 4.23PGRR43 pKa = 11.84YY44 pKa = 8.22RR45 pKa = 11.84VVTLCNICHH54 pKa = 6.02SSLRR58 pKa = 11.84LFVEE62 pKa = 4.47VADD65 pKa = 4.31EE66 pKa = 4.03SLIRR70 pKa = 11.84LFQQLLLDD78 pKa = 4.0GLGIICATCHH88 pKa = 6.56KK89 pKa = 10.12EE90 pKa = 3.8HH91 pKa = 6.88FSDD94 pKa = 3.74GRR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 3.42
Molecular weight: 11.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q647H9|Q647H9_9PAPI Replication protein E1 OS=Canis familiaris papillomavirus 2 OX=292792 GN=E1 PE=3 SV=1
MM1 pKa = 7.56LEE3 pKa = 3.76SLARR7 pKa = 11.84RR8 pKa = 11.84FDD10 pKa = 3.61VLQEE14 pKa = 4.12VMLHH18 pKa = 6.22HH19 pKa = 6.59YY20 pKa = 10.23EE21 pKa = 4.25SGSKK25 pKa = 10.55KK26 pKa = 9.87LTDD29 pKa = 3.21QCNYY33 pKa = 8.66WEE35 pKa = 4.16AARR38 pKa = 11.84RR39 pKa = 11.84EE40 pKa = 4.35SALLHH45 pKa = 5.18YY46 pKa = 10.22ARR48 pKa = 11.84KK49 pKa = 9.78NRR51 pKa = 11.84ITRR54 pKa = 11.84LGITPVPSLATSEE67 pKa = 4.31NSAKK71 pKa = 10.39KK72 pKa = 10.06AIRR75 pKa = 11.84MSLLLSSLCNSPYY88 pKa = 10.24KK89 pKa = 10.47DD90 pKa = 4.25EE91 pKa = 4.64PWTLADD97 pKa = 3.56TSIEE101 pKa = 3.93LWEE104 pKa = 5.01CPPKK108 pKa = 11.06GCFKK112 pKa = 10.9KK113 pKa = 10.85GGFTVDD119 pKa = 3.23VLFDD123 pKa = 4.04NDD125 pKa = 3.83PGNVFPYY132 pKa = 9.12TAWAYY137 pKa = 9.81IYY139 pKa = 9.67YY140 pKa = 10.24QNQNDD145 pKa = 3.52EE146 pKa = 4.5WIKK149 pKa = 10.83VPGQVDD155 pKa = 3.92YY156 pKa = 11.68DD157 pKa = 3.74GLFYY161 pKa = 10.68TDD163 pKa = 3.45EE164 pKa = 5.82DD165 pKa = 4.25GEE167 pKa = 4.14KK168 pKa = 10.0RR169 pKa = 11.84YY170 pKa = 10.12YY171 pKa = 8.82QTFAKK176 pKa = 10.41DD177 pKa = 3.22ALRR180 pKa = 11.84FGTTGMWKK188 pKa = 9.19VCYY191 pKa = 9.32KK192 pKa = 10.5HH193 pKa = 6.57HH194 pKa = 6.45VLSALISSSGSSNSAASSSWQSNIEE219 pKa = 4.16SSSGDD224 pKa = 3.02QWSAQWQWEE233 pKa = 4.62GPTGSSASSTAASVSSSSTCSRR255 pKa = 11.84TGGTRR260 pKa = 11.84EE261 pKa = 3.92AGGSFDD267 pKa = 4.87QGGSSQAGGAEE278 pKa = 4.04RR279 pKa = 11.84GRR281 pKa = 11.84GRR283 pKa = 11.84GRR285 pKa = 11.84GRR287 pKa = 11.84GRR289 pKa = 11.84GRR291 pKa = 11.84GRR293 pKa = 11.84GRR295 pKa = 11.84ASSGSGSGSGSTSRR309 pKa = 11.84SGSRR313 pKa = 11.84SDD315 pKa = 3.56SVSGSGGAKK324 pKa = 9.08PVYY327 pKa = 8.28PQPRR331 pKa = 11.84VGGNKK336 pKa = 9.67APQKK340 pKa = 9.52RR341 pKa = 11.84QRR343 pKa = 11.84EE344 pKa = 4.33APSDD348 pKa = 3.87TEE350 pKa = 3.9DD351 pKa = 3.22LPRR354 pKa = 11.84NRR356 pKa = 11.84GVLGRR361 pKa = 11.84GRR363 pKa = 11.84GASPSTWSSNVRR375 pKa = 11.84NIPPNTPTIPTLSTPGPCPGDD396 pKa = 3.15RR397 pKa = 11.84AGVGRR402 pKa = 11.84GGGPRR407 pKa = 11.84EE408 pKa = 3.99PKK410 pKa = 10.49APRR413 pKa = 11.84LSPAVPLGTVGRR425 pKa = 11.84RR426 pKa = 11.84PGEE429 pKa = 3.84PQEE432 pKa = 4.16PLEE435 pKa = 4.39PEE437 pKa = 4.05PGRR440 pKa = 11.84SQEE443 pKa = 3.92QNRR446 pKa = 11.84PFPVILVKK454 pKa = 10.93GDD456 pKa = 3.54GNASKK461 pKa = 10.44CWRR464 pKa = 11.84RR465 pKa = 11.84RR466 pKa = 11.84LRR468 pKa = 11.84LRR470 pKa = 11.84YY471 pKa = 9.49SGLFEE476 pKa = 4.86SISTGFSWTTASGPHH491 pKa = 5.7RR492 pKa = 11.84VCRR495 pKa = 11.84PRR497 pKa = 11.84MLIAFNSDD505 pKa = 2.84GQRR508 pKa = 11.84DD509 pKa = 3.79HH510 pKa = 6.78FLKK513 pKa = 10.7NIKK516 pKa = 9.37IPKK519 pKa = 9.25VLDD522 pKa = 3.12IALGSFDD529 pKa = 4.16SLL531 pKa = 4.03
MM1 pKa = 7.56LEE3 pKa = 3.76SLARR7 pKa = 11.84RR8 pKa = 11.84FDD10 pKa = 3.61VLQEE14 pKa = 4.12VMLHH18 pKa = 6.22HH19 pKa = 6.59YY20 pKa = 10.23EE21 pKa = 4.25SGSKK25 pKa = 10.55KK26 pKa = 9.87LTDD29 pKa = 3.21QCNYY33 pKa = 8.66WEE35 pKa = 4.16AARR38 pKa = 11.84RR39 pKa = 11.84EE40 pKa = 4.35SALLHH45 pKa = 5.18YY46 pKa = 10.22ARR48 pKa = 11.84KK49 pKa = 9.78NRR51 pKa = 11.84ITRR54 pKa = 11.84LGITPVPSLATSEE67 pKa = 4.31NSAKK71 pKa = 10.39KK72 pKa = 10.06AIRR75 pKa = 11.84MSLLLSSLCNSPYY88 pKa = 10.24KK89 pKa = 10.47DD90 pKa = 4.25EE91 pKa = 4.64PWTLADD97 pKa = 3.56TSIEE101 pKa = 3.93LWEE104 pKa = 5.01CPPKK108 pKa = 11.06GCFKK112 pKa = 10.9KK113 pKa = 10.85GGFTVDD119 pKa = 3.23VLFDD123 pKa = 4.04NDD125 pKa = 3.83PGNVFPYY132 pKa = 9.12TAWAYY137 pKa = 9.81IYY139 pKa = 9.67YY140 pKa = 10.24QNQNDD145 pKa = 3.52EE146 pKa = 4.5WIKK149 pKa = 10.83VPGQVDD155 pKa = 3.92YY156 pKa = 11.68DD157 pKa = 3.74GLFYY161 pKa = 10.68TDD163 pKa = 3.45EE164 pKa = 5.82DD165 pKa = 4.25GEE167 pKa = 4.14KK168 pKa = 10.0RR169 pKa = 11.84YY170 pKa = 10.12YY171 pKa = 8.82QTFAKK176 pKa = 10.41DD177 pKa = 3.22ALRR180 pKa = 11.84FGTTGMWKK188 pKa = 9.19VCYY191 pKa = 9.32KK192 pKa = 10.5HH193 pKa = 6.57HH194 pKa = 6.45VLSALISSSGSSNSAASSSWQSNIEE219 pKa = 4.16SSSGDD224 pKa = 3.02QWSAQWQWEE233 pKa = 4.62GPTGSSASSTAASVSSSSTCSRR255 pKa = 11.84TGGTRR260 pKa = 11.84EE261 pKa = 3.92AGGSFDD267 pKa = 4.87QGGSSQAGGAEE278 pKa = 4.04RR279 pKa = 11.84GRR281 pKa = 11.84GRR283 pKa = 11.84GRR285 pKa = 11.84GRR287 pKa = 11.84GRR289 pKa = 11.84GRR291 pKa = 11.84GRR293 pKa = 11.84GRR295 pKa = 11.84ASSGSGSGSGSTSRR309 pKa = 11.84SGSRR313 pKa = 11.84SDD315 pKa = 3.56SVSGSGGAKK324 pKa = 9.08PVYY327 pKa = 8.28PQPRR331 pKa = 11.84VGGNKK336 pKa = 9.67APQKK340 pKa = 9.52RR341 pKa = 11.84QRR343 pKa = 11.84EE344 pKa = 4.33APSDD348 pKa = 3.87TEE350 pKa = 3.9DD351 pKa = 3.22LPRR354 pKa = 11.84NRR356 pKa = 11.84GVLGRR361 pKa = 11.84GRR363 pKa = 11.84GASPSTWSSNVRR375 pKa = 11.84NIPPNTPTIPTLSTPGPCPGDD396 pKa = 3.15RR397 pKa = 11.84AGVGRR402 pKa = 11.84GGGPRR407 pKa = 11.84EE408 pKa = 3.99PKK410 pKa = 10.49APRR413 pKa = 11.84LSPAVPLGTVGRR425 pKa = 11.84RR426 pKa = 11.84PGEE429 pKa = 3.84PQEE432 pKa = 4.16PLEE435 pKa = 4.39PEE437 pKa = 4.05PGRR440 pKa = 11.84SQEE443 pKa = 3.92QNRR446 pKa = 11.84PFPVILVKK454 pKa = 10.93GDD456 pKa = 3.54GNASKK461 pKa = 10.44CWRR464 pKa = 11.84RR465 pKa = 11.84RR466 pKa = 11.84LRR468 pKa = 11.84LRR470 pKa = 11.84YY471 pKa = 9.49SGLFEE476 pKa = 4.86SISTGFSWTTASGPHH491 pKa = 5.7RR492 pKa = 11.84VCRR495 pKa = 11.84PRR497 pKa = 11.84MLIAFNSDD505 pKa = 2.84GQRR508 pKa = 11.84DD509 pKa = 3.79HH510 pKa = 6.78FLKK513 pKa = 10.7NIKK516 pKa = 9.37IPKK519 pKa = 9.25VLDD522 pKa = 3.12IALGSFDD529 pKa = 4.16SLL531 pKa = 4.03
Molecular weight: 57.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2754 |
41 |
607 |
344.3 |
38.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.483 ± 0.668 | 2.687 ± 0.99 |
5.556 ± 0.372 | 6.173 ± 0.51 |
4.357 ± 0.571 | 8.315 ± 1.132 |
1.961 ± 0.327 | 4.103 ± 0.637 |
4.466 ± 0.648 | 9.005 ± 0.912 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.743 ± 0.423 | 4.031 ± 0.6 |
7.734 ± 1.497 | 3.595 ± 0.326 |
6.572 ± 0.816 | 8.533 ± 1.247 |
5.81 ± 0.844 | 5.846 ± 0.767 |
1.38 ± 0.36 | 2.651 ± 0.408 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
