
Bos taurus papillomavirus 21
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyoxipapillomavirus; Dyoxipapillomavirus 2
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
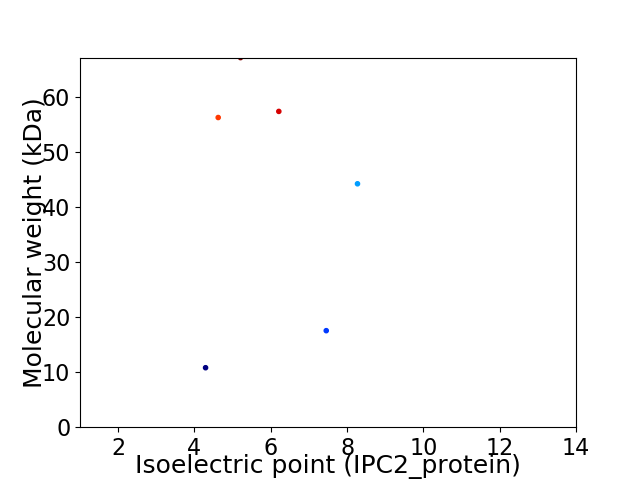
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B2K2D0|A0A1B2K2D0_9PAPI Minor capsid protein L2 OS=Bos taurus papillomavirus 21 OX=1887219 GN=L2 PE=3 SV=1
MM1 pKa = 7.58MIGEE5 pKa = 4.58TPTIRR10 pKa = 11.84DD11 pKa = 2.91IDD13 pKa = 4.17LDD15 pKa = 3.98LTEE18 pKa = 6.36LIDD21 pKa = 4.21PQNLLCEE28 pKa = 4.22EE29 pKa = 4.68EE30 pKa = 4.13ILAEE34 pKa = 4.01EE35 pKa = 4.2VLEE38 pKa = 4.9EE39 pKa = 4.06EE40 pKa = 4.47LCPYY44 pKa = 10.38SLRR47 pKa = 11.84TCCASCSALLRR58 pKa = 11.84IFVVTSSDD66 pKa = 4.11SIRR69 pKa = 11.84TLHH72 pKa = 6.66QLLLSDD78 pKa = 5.59LSFLCIPCSQRR89 pKa = 11.84NLRR92 pKa = 11.84NEE94 pKa = 3.91RR95 pKa = 3.56
MM1 pKa = 7.58MIGEE5 pKa = 4.58TPTIRR10 pKa = 11.84DD11 pKa = 2.91IDD13 pKa = 4.17LDD15 pKa = 3.98LTEE18 pKa = 6.36LIDD21 pKa = 4.21PQNLLCEE28 pKa = 4.22EE29 pKa = 4.68EE30 pKa = 4.13ILAEE34 pKa = 4.01EE35 pKa = 4.2VLEE38 pKa = 4.9EE39 pKa = 4.06EE40 pKa = 4.47LCPYY44 pKa = 10.38SLRR47 pKa = 11.84TCCASCSALLRR58 pKa = 11.84IFVVTSSDD66 pKa = 4.11SIRR69 pKa = 11.84TLHH72 pKa = 6.66QLLLSDD78 pKa = 5.59LSFLCIPCSQRR89 pKa = 11.84NLRR92 pKa = 11.84NEE94 pKa = 3.91RR95 pKa = 3.56
Molecular weight: 10.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B2K226|A0A1B2K226_9PAPI Major capsid protein L1 OS=Bos taurus papillomavirus 21 OX=1887219 GN=L1 PE=3 SV=1
MM1 pKa = 7.64EE2 pKa = 4.81SLADD6 pKa = 3.63RR7 pKa = 11.84FEE9 pKa = 4.96EE10 pKa = 4.15IQEE13 pKa = 4.22LLLDD17 pKa = 4.66LYY19 pKa = 10.9EE20 pKa = 4.6AGKK23 pKa = 10.47RR24 pKa = 11.84DD25 pKa = 3.47IQSQIIHH32 pKa = 6.71WDD34 pKa = 3.52YY35 pKa = 11.29LRR37 pKa = 11.84KK38 pKa = 9.96EE39 pKa = 4.32SVLLYY44 pKa = 9.19YY45 pKa = 10.58ARR47 pKa = 11.84QRR49 pKa = 11.84GLTRR53 pKa = 11.84LGLNTVPVLQASEE66 pKa = 3.94IRR68 pKa = 11.84AKK70 pKa = 10.18NAIMMGIILRR80 pKa = 11.84SLANSEE86 pKa = 4.37FGGEE90 pKa = 4.1DD91 pKa = 2.71WTLQDD96 pKa = 4.19TSIEE100 pKa = 4.28MYY102 pKa = 10.13RR103 pKa = 11.84APPQDD108 pKa = 3.22TFKK111 pKa = 11.04KK112 pKa = 9.27QGSPVEE118 pKa = 4.16VMFDD122 pKa = 3.52GDD124 pKa = 3.84PEE126 pKa = 4.55NINVYY131 pKa = 7.61TNWGRR136 pKa = 11.84IYY138 pKa = 11.06YY139 pKa = 9.91QDD141 pKa = 3.66TEE143 pKa = 4.55NNWRR147 pKa = 11.84VAEE150 pKa = 4.34GQVSYY155 pKa = 11.4DD156 pKa = 2.88GLYY159 pKa = 9.66YY160 pKa = 10.22DD161 pKa = 4.0TVNGQRR167 pKa = 11.84VFFVKK172 pKa = 10.34FDD174 pKa = 3.47EE175 pKa = 4.32QARR178 pKa = 11.84TFSKK182 pKa = 9.36TGSWRR187 pKa = 11.84VKK189 pKa = 9.69YY190 pKa = 10.21KK191 pKa = 11.0NKK193 pKa = 9.77FVSEE197 pKa = 4.55SVSSSSTPVSGSSQEE212 pKa = 3.91RR213 pKa = 11.84PRR215 pKa = 11.84VSFDD219 pKa = 3.14TDD221 pKa = 3.43SEE223 pKa = 4.38EE224 pKa = 4.88EE225 pKa = 4.21EE226 pKa = 4.69GDD228 pKa = 3.22HH229 pKa = 6.91HH230 pKa = 7.26RR231 pKa = 11.84RR232 pKa = 11.84DD233 pKa = 3.58PRR235 pKa = 11.84HH236 pKa = 6.05PLSAGRR242 pKa = 11.84SRR244 pKa = 11.84SRR246 pKa = 11.84SRR248 pKa = 11.84VSRR251 pKa = 11.84PEE253 pKa = 3.62PEE255 pKa = 3.81AGGRR259 pKa = 11.84GRR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84QGEE268 pKa = 3.91GAEE271 pKa = 4.36DD272 pKa = 3.78FGLVPEE278 pKa = 4.71EE279 pKa = 4.19VGSGHH284 pKa = 5.69RR285 pKa = 11.84TVPKK289 pKa = 10.28NIRR292 pKa = 11.84GRR294 pKa = 11.84LARR297 pKa = 11.84LQAEE301 pKa = 4.34AADD304 pKa = 3.88PPVILVRR311 pKa = 11.84GSINSLKK318 pKa = 10.24SWRR321 pKa = 11.84HH322 pKa = 4.3RR323 pKa = 11.84CNTKK327 pKa = 9.93HH328 pKa = 6.26RR329 pKa = 11.84CLFQEE334 pKa = 3.92IGTTFVWSSKK344 pKa = 9.36DD345 pKa = 3.23TSRR348 pKa = 11.84DD349 pKa = 2.9RR350 pKa = 11.84GRR352 pKa = 11.84VVVAFRR358 pKa = 11.84DD359 pKa = 4.1EE360 pKa = 4.27VQLTEE365 pKa = 4.21FLRR368 pKa = 11.84VVPMPKK374 pKa = 9.28SCSFARR380 pKa = 11.84GHH382 pKa = 6.95LDD384 pKa = 3.22SLL386 pKa = 4.26
MM1 pKa = 7.64EE2 pKa = 4.81SLADD6 pKa = 3.63RR7 pKa = 11.84FEE9 pKa = 4.96EE10 pKa = 4.15IQEE13 pKa = 4.22LLLDD17 pKa = 4.66LYY19 pKa = 10.9EE20 pKa = 4.6AGKK23 pKa = 10.47RR24 pKa = 11.84DD25 pKa = 3.47IQSQIIHH32 pKa = 6.71WDD34 pKa = 3.52YY35 pKa = 11.29LRR37 pKa = 11.84KK38 pKa = 9.96EE39 pKa = 4.32SVLLYY44 pKa = 9.19YY45 pKa = 10.58ARR47 pKa = 11.84QRR49 pKa = 11.84GLTRR53 pKa = 11.84LGLNTVPVLQASEE66 pKa = 3.94IRR68 pKa = 11.84AKK70 pKa = 10.18NAIMMGIILRR80 pKa = 11.84SLANSEE86 pKa = 4.37FGGEE90 pKa = 4.1DD91 pKa = 2.71WTLQDD96 pKa = 4.19TSIEE100 pKa = 4.28MYY102 pKa = 10.13RR103 pKa = 11.84APPQDD108 pKa = 3.22TFKK111 pKa = 11.04KK112 pKa = 9.27QGSPVEE118 pKa = 4.16VMFDD122 pKa = 3.52GDD124 pKa = 3.84PEE126 pKa = 4.55NINVYY131 pKa = 7.61TNWGRR136 pKa = 11.84IYY138 pKa = 11.06YY139 pKa = 9.91QDD141 pKa = 3.66TEE143 pKa = 4.55NNWRR147 pKa = 11.84VAEE150 pKa = 4.34GQVSYY155 pKa = 11.4DD156 pKa = 2.88GLYY159 pKa = 9.66YY160 pKa = 10.22DD161 pKa = 4.0TVNGQRR167 pKa = 11.84VFFVKK172 pKa = 10.34FDD174 pKa = 3.47EE175 pKa = 4.32QARR178 pKa = 11.84TFSKK182 pKa = 9.36TGSWRR187 pKa = 11.84VKK189 pKa = 9.69YY190 pKa = 10.21KK191 pKa = 11.0NKK193 pKa = 9.77FVSEE197 pKa = 4.55SVSSSSTPVSGSSQEE212 pKa = 3.91RR213 pKa = 11.84PRR215 pKa = 11.84VSFDD219 pKa = 3.14TDD221 pKa = 3.43SEE223 pKa = 4.38EE224 pKa = 4.88EE225 pKa = 4.21EE226 pKa = 4.69GDD228 pKa = 3.22HH229 pKa = 6.91HH230 pKa = 7.26RR231 pKa = 11.84RR232 pKa = 11.84DD233 pKa = 3.58PRR235 pKa = 11.84HH236 pKa = 6.05PLSAGRR242 pKa = 11.84SRR244 pKa = 11.84SRR246 pKa = 11.84SRR248 pKa = 11.84VSRR251 pKa = 11.84PEE253 pKa = 3.62PEE255 pKa = 3.81AGGRR259 pKa = 11.84GRR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84QGEE268 pKa = 3.91GAEE271 pKa = 4.36DD272 pKa = 3.78FGLVPEE278 pKa = 4.71EE279 pKa = 4.19VGSGHH284 pKa = 5.69RR285 pKa = 11.84TVPKK289 pKa = 10.28NIRR292 pKa = 11.84GRR294 pKa = 11.84LARR297 pKa = 11.84LQAEE301 pKa = 4.34AADD304 pKa = 3.88PPVILVRR311 pKa = 11.84GSINSLKK318 pKa = 10.24SWRR321 pKa = 11.84HH322 pKa = 4.3RR323 pKa = 11.84CNTKK327 pKa = 9.93HH328 pKa = 6.26RR329 pKa = 11.84CLFQEE334 pKa = 3.92IGTTFVWSSKK344 pKa = 9.36DD345 pKa = 3.23TSRR348 pKa = 11.84DD349 pKa = 2.9RR350 pKa = 11.84GRR352 pKa = 11.84VVVAFRR358 pKa = 11.84DD359 pKa = 4.1EE360 pKa = 4.27VQLTEE365 pKa = 4.21FLRR368 pKa = 11.84VVPMPKK374 pKa = 9.28SCSFARR380 pKa = 11.84GHH382 pKa = 6.95LDD384 pKa = 3.22SLL386 pKa = 4.26
Molecular weight: 44.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2239 |
95 |
590 |
373.2 |
42.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.449 ± 0.466 | 2.188 ± 0.813 |
6.699 ± 0.332 | 6.878 ± 0.347 |
4.69 ± 0.517 | 6.208 ± 0.813 |
1.831 ± 0.226 | 5.181 ± 0.445 |
4.868 ± 1.062 | 8.709 ± 0.969 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.697 ± 0.302 | 4.6 ± 0.518 |
5.538 ± 1.034 | 3.93 ± 0.427 |
6.789 ± 1.216 | 7.057 ± 0.801 |
6.699 ± 0.545 | 6.342 ± 0.723 |
1.251 ± 0.343 | 3.394 ± 0.392 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
