
Acidovorax phage ACP17
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Busanvirus; Acidovorax virus ACP17
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
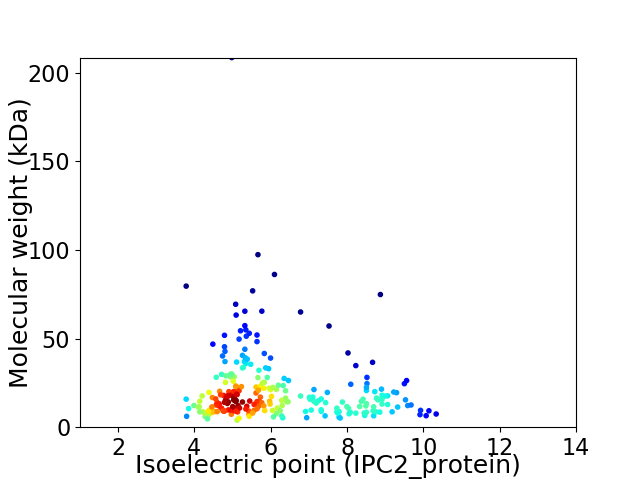
Virtual 2D-PAGE plot for 253 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A218M2T3|A0A218M2T3_9CAUD Uncharacterized protein OS=Acidovorax phage ACP17 OX=2010329 PE=4 SV=1
MM1 pKa = 7.57SSNNGASTILGITAGLGSVDD21 pKa = 4.18FVFDD25 pKa = 3.98KK26 pKa = 10.92KK27 pKa = 10.82ALQIVPPVDD36 pKa = 4.68FIFGADD42 pKa = 3.01KK43 pKa = 11.01CFPDD47 pKa = 4.43WMDD50 pKa = 3.11QSGGSIIVDD59 pKa = 2.8IDD61 pKa = 3.77GCCFDD66 pKa = 4.81VANDD70 pKa = 3.8WVSVAGDD77 pKa = 4.41SIVLDD82 pKa = 3.98ATKK85 pKa = 10.43GWQDD89 pKa = 2.93ASGDD93 pKa = 3.8QLVLDD98 pKa = 4.1NGKK101 pKa = 9.71SAVCPPDD108 pKa = 3.4WEE110 pKa = 4.36PQSGGSIDD118 pKa = 3.48VDD120 pKa = 4.73FYY122 pKa = 11.72CCTTGGGEE130 pKa = 4.68ALPIDD135 pKa = 4.5LGAVRR140 pKa = 11.84FTEE143 pKa = 4.38GSVFVADD150 pKa = 5.75AITDD154 pKa = 3.64PRR156 pKa = 11.84ISLGFYY162 pKa = 10.12EE163 pKa = 4.99GATFEE168 pKa = 4.62ADD170 pKa = 3.25LVRR173 pKa = 11.84YY174 pKa = 8.84PSVLLSADD182 pKa = 3.46AVGGEE187 pKa = 4.28YY188 pKa = 10.63VEE190 pKa = 4.35ASIYY194 pKa = 10.01TDD196 pKa = 3.18VVLPADD202 pKa = 3.94AAAGEE207 pKa = 4.16YY208 pKa = 10.83LEE210 pKa = 5.41LALAVFPAIDD220 pKa = 3.88LQAVAVDD227 pKa = 4.36GASLEE232 pKa = 4.25AKK234 pKa = 10.51VEE236 pKa = 4.23FTTGFPQTYY245 pKa = 8.07TQGEE249 pKa = 4.3WAVADD254 pKa = 4.42LEE256 pKa = 4.31ITPYY260 pKa = 10.72EE261 pKa = 4.0GLIPVAHH268 pKa = 6.46TGEE271 pKa = 4.15VLQVSVEE278 pKa = 4.43TITGFQVPVYY288 pKa = 9.99VGEE291 pKa = 4.26SADD294 pKa = 3.57ASLASTTTFDD304 pKa = 4.12ANAHH308 pKa = 5.27TGEE311 pKa = 4.22NASADD316 pKa = 3.93LAVAPPAYY324 pKa = 9.21LTAEE328 pKa = 4.43LATGEE333 pKa = 4.42SFVASLALGASSFGVNFAHH352 pKa = 6.96EE353 pKa = 4.43GSEE356 pKa = 4.48AYY358 pKa = 10.49VSLDD362 pKa = 3.28TRR364 pKa = 11.84PAADD368 pKa = 5.1LFWQFYY374 pKa = 10.29AGEE377 pKa = 4.2QGSVTLSLGANLGEE391 pKa = 4.06VGFFEE396 pKa = 4.82GVRR399 pKa = 11.84AEE401 pKa = 4.02ASLGEE406 pKa = 4.33EE407 pKa = 3.91PNVYY411 pKa = 10.23AFHH414 pKa = 6.89GEE416 pKa = 4.07VMDD419 pKa = 3.77WALRR423 pKa = 11.84TEE425 pKa = 4.92DD426 pKa = 3.64GFQAGVSQGEE436 pKa = 4.5SASLEE441 pKa = 4.09LNVRR445 pKa = 11.84VSEE448 pKa = 4.26GLGEE452 pKa = 3.99LAAADD457 pKa = 4.08GSTVFANLGALKK469 pKa = 10.25AIPLAVTFYY478 pKa = 11.29NSYY481 pKa = 8.82TVHH484 pKa = 7.16SGIGASTFLDD494 pKa = 3.96LDD496 pKa = 3.88SGCASGHH503 pKa = 5.8NPGKK507 pKa = 10.28QDD509 pKa = 2.82IHH511 pKa = 8.1RR512 pKa = 11.84IEE514 pKa = 4.26MNDD517 pKa = 3.98AEE519 pKa = 5.14FPDD522 pKa = 3.59QRR524 pKa = 11.84FDD526 pKa = 3.76GDD528 pKa = 3.93RR529 pKa = 11.84TCVSLDD535 pKa = 3.7LSAQPRR541 pKa = 11.84FSFNFFGGEE550 pKa = 3.94TFRR553 pKa = 11.84TVDD556 pKa = 3.25AADD559 pKa = 3.74PSLRR563 pKa = 11.84TVLSNEE569 pKa = 3.75GATMRR574 pKa = 11.84FALEE578 pKa = 3.82SNINFRR584 pKa = 11.84LCPGNFIPDD593 pKa = 3.41GDD595 pKa = 3.91NVYY598 pKa = 10.77VEE600 pKa = 5.29LDD602 pKa = 3.55VEE604 pKa = 5.06DD605 pKa = 4.23VGNCQSDD612 pKa = 4.36YY613 pKa = 11.34AFHH616 pKa = 6.53GVNVSAWLQANQNFQPVGHH635 pKa = 6.7TGEE638 pKa = 4.37TLTFDD643 pKa = 3.69LTTAEE648 pKa = 4.26LWTLVGQAGEE658 pKa = 4.35TFSMASQPDD667 pKa = 3.99PQPRR671 pKa = 11.84AYY673 pKa = 10.18EE674 pKa = 4.24GVRR677 pKa = 11.84VDD679 pKa = 3.21ATFYY683 pKa = 10.37EE684 pKa = 4.47PPIVGAEE691 pKa = 4.06GATAEE696 pKa = 4.07ASLTIKK702 pKa = 11.08YY703 pKa = 8.09EE704 pKa = 4.1VKK706 pKa = 10.27FDD708 pKa = 4.45EE709 pKa = 5.24VGCLDD714 pKa = 5.71NEE716 pKa = 5.3YY717 pKa = 10.06IWQDD721 pKa = 3.58DD722 pKa = 3.74NGDD725 pKa = 4.5PITEE729 pKa = 4.5LFNPVAVEE737 pKa = 3.94MDD739 pKa = 4.13PFSHH743 pKa = 8.03DD744 pKa = 2.9IRR746 pKa = 11.84AHH748 pKa = 5.96CEE750 pKa = 3.48
MM1 pKa = 7.57SSNNGASTILGITAGLGSVDD21 pKa = 4.18FVFDD25 pKa = 3.98KK26 pKa = 10.92KK27 pKa = 10.82ALQIVPPVDD36 pKa = 4.68FIFGADD42 pKa = 3.01KK43 pKa = 11.01CFPDD47 pKa = 4.43WMDD50 pKa = 3.11QSGGSIIVDD59 pKa = 2.8IDD61 pKa = 3.77GCCFDD66 pKa = 4.81VANDD70 pKa = 3.8WVSVAGDD77 pKa = 4.41SIVLDD82 pKa = 3.98ATKK85 pKa = 10.43GWQDD89 pKa = 2.93ASGDD93 pKa = 3.8QLVLDD98 pKa = 4.1NGKK101 pKa = 9.71SAVCPPDD108 pKa = 3.4WEE110 pKa = 4.36PQSGGSIDD118 pKa = 3.48VDD120 pKa = 4.73FYY122 pKa = 11.72CCTTGGGEE130 pKa = 4.68ALPIDD135 pKa = 4.5LGAVRR140 pKa = 11.84FTEE143 pKa = 4.38GSVFVADD150 pKa = 5.75AITDD154 pKa = 3.64PRR156 pKa = 11.84ISLGFYY162 pKa = 10.12EE163 pKa = 4.99GATFEE168 pKa = 4.62ADD170 pKa = 3.25LVRR173 pKa = 11.84YY174 pKa = 8.84PSVLLSADD182 pKa = 3.46AVGGEE187 pKa = 4.28YY188 pKa = 10.63VEE190 pKa = 4.35ASIYY194 pKa = 10.01TDD196 pKa = 3.18VVLPADD202 pKa = 3.94AAAGEE207 pKa = 4.16YY208 pKa = 10.83LEE210 pKa = 5.41LALAVFPAIDD220 pKa = 3.88LQAVAVDD227 pKa = 4.36GASLEE232 pKa = 4.25AKK234 pKa = 10.51VEE236 pKa = 4.23FTTGFPQTYY245 pKa = 8.07TQGEE249 pKa = 4.3WAVADD254 pKa = 4.42LEE256 pKa = 4.31ITPYY260 pKa = 10.72EE261 pKa = 4.0GLIPVAHH268 pKa = 6.46TGEE271 pKa = 4.15VLQVSVEE278 pKa = 4.43TITGFQVPVYY288 pKa = 9.99VGEE291 pKa = 4.26SADD294 pKa = 3.57ASLASTTTFDD304 pKa = 4.12ANAHH308 pKa = 5.27TGEE311 pKa = 4.22NASADD316 pKa = 3.93LAVAPPAYY324 pKa = 9.21LTAEE328 pKa = 4.43LATGEE333 pKa = 4.42SFVASLALGASSFGVNFAHH352 pKa = 6.96EE353 pKa = 4.43GSEE356 pKa = 4.48AYY358 pKa = 10.49VSLDD362 pKa = 3.28TRR364 pKa = 11.84PAADD368 pKa = 5.1LFWQFYY374 pKa = 10.29AGEE377 pKa = 4.2QGSVTLSLGANLGEE391 pKa = 4.06VGFFEE396 pKa = 4.82GVRR399 pKa = 11.84AEE401 pKa = 4.02ASLGEE406 pKa = 4.33EE407 pKa = 3.91PNVYY411 pKa = 10.23AFHH414 pKa = 6.89GEE416 pKa = 4.07VMDD419 pKa = 3.77WALRR423 pKa = 11.84TEE425 pKa = 4.92DD426 pKa = 3.64GFQAGVSQGEE436 pKa = 4.5SASLEE441 pKa = 4.09LNVRR445 pKa = 11.84VSEE448 pKa = 4.26GLGEE452 pKa = 3.99LAAADD457 pKa = 4.08GSTVFANLGALKK469 pKa = 10.25AIPLAVTFYY478 pKa = 11.29NSYY481 pKa = 8.82TVHH484 pKa = 7.16SGIGASTFLDD494 pKa = 3.96LDD496 pKa = 3.88SGCASGHH503 pKa = 5.8NPGKK507 pKa = 10.28QDD509 pKa = 2.82IHH511 pKa = 8.1RR512 pKa = 11.84IEE514 pKa = 4.26MNDD517 pKa = 3.98AEE519 pKa = 5.14FPDD522 pKa = 3.59QRR524 pKa = 11.84FDD526 pKa = 3.76GDD528 pKa = 3.93RR529 pKa = 11.84TCVSLDD535 pKa = 3.7LSAQPRR541 pKa = 11.84FSFNFFGGEE550 pKa = 3.94TFRR553 pKa = 11.84TVDD556 pKa = 3.25AADD559 pKa = 3.74PSLRR563 pKa = 11.84TVLSNEE569 pKa = 3.75GATMRR574 pKa = 11.84FALEE578 pKa = 3.82SNINFRR584 pKa = 11.84LCPGNFIPDD593 pKa = 3.41GDD595 pKa = 3.91NVYY598 pKa = 10.77VEE600 pKa = 5.29LDD602 pKa = 3.55VEE604 pKa = 5.06DD605 pKa = 4.23VGNCQSDD612 pKa = 4.36YY613 pKa = 11.34AFHH616 pKa = 6.53GVNVSAWLQANQNFQPVGHH635 pKa = 6.7TGEE638 pKa = 4.37TLTFDD643 pKa = 3.69LTTAEE648 pKa = 4.26LWTLVGQAGEE658 pKa = 4.35TFSMASQPDD667 pKa = 3.99PQPRR671 pKa = 11.84AYY673 pKa = 10.18EE674 pKa = 4.24GVRR677 pKa = 11.84VDD679 pKa = 3.21ATFYY683 pKa = 10.37EE684 pKa = 4.47PPIVGAEE691 pKa = 4.06GATAEE696 pKa = 4.07ASLTIKK702 pKa = 11.08YY703 pKa = 8.09EE704 pKa = 4.1VKK706 pKa = 10.27FDD708 pKa = 4.45EE709 pKa = 5.24VGCLDD714 pKa = 5.71NEE716 pKa = 5.3YY717 pKa = 10.06IWQDD721 pKa = 3.58DD722 pKa = 3.74NGDD725 pKa = 4.5PITEE729 pKa = 4.5LFNPVAVEE737 pKa = 3.94MDD739 pKa = 4.13PFSHH743 pKa = 8.03DD744 pKa = 2.9IRR746 pKa = 11.84AHH748 pKa = 5.96CEE750 pKa = 3.48
Molecular weight: 79.56 kDa
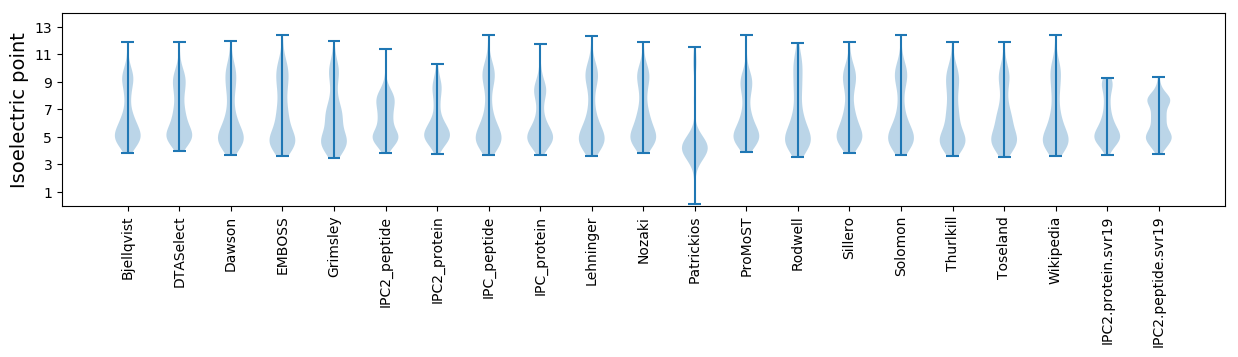
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A218M3A9|A0A218M3A9_9CAUD Uncharacterized protein OS=Acidovorax phage ACP17 OX=2010329 PE=4 SV=1
MM1 pKa = 7.92AKK3 pKa = 9.03FTSLNTQVYY12 pKa = 9.79IPDD15 pKa = 3.46NFAPLGKK22 pKa = 9.59EE23 pKa = 3.55KK24 pKa = 10.5KK25 pKa = 10.56GYY27 pKa = 10.12FVDD30 pKa = 3.83LGDD33 pKa = 4.05RR34 pKa = 11.84SVHH37 pKa = 5.82SIHH40 pKa = 7.58RR41 pKa = 11.84GGCLKK46 pKa = 10.82ALTPYY51 pKa = 9.96KK52 pKa = 10.38GRR54 pKa = 11.84YY55 pKa = 7.94KK56 pKa = 10.54LRR58 pKa = 11.84PAAGGWLARR67 pKa = 11.84DD68 pKa = 4.4VIAKK72 pKa = 9.94AKK74 pKa = 8.4RR75 pKa = 11.84TSKK78 pKa = 9.17TPRR81 pKa = 11.84FARR84 pKa = 11.84VV85 pKa = 2.74
MM1 pKa = 7.92AKK3 pKa = 9.03FTSLNTQVYY12 pKa = 9.79IPDD15 pKa = 3.46NFAPLGKK22 pKa = 9.59EE23 pKa = 3.55KK24 pKa = 10.5KK25 pKa = 10.56GYY27 pKa = 10.12FVDD30 pKa = 3.83LGDD33 pKa = 4.05RR34 pKa = 11.84SVHH37 pKa = 5.82SIHH40 pKa = 7.58RR41 pKa = 11.84GGCLKK46 pKa = 10.82ALTPYY51 pKa = 9.96KK52 pKa = 10.38GRR54 pKa = 11.84YY55 pKa = 7.94KK56 pKa = 10.54LRR58 pKa = 11.84PAAGGWLARR67 pKa = 11.84DD68 pKa = 4.4VIAKK72 pKa = 9.94AKK74 pKa = 8.4RR75 pKa = 11.84TSKK78 pKa = 9.17TPRR81 pKa = 11.84FARR84 pKa = 11.84VV85 pKa = 2.74
Molecular weight: 9.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
48500 |
40 |
1903 |
191.7 |
21.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.532 ± 0.241 | 1.157 ± 0.072 |
6.161 ± 0.13 | 6.66 ± 0.168 |
4.365 ± 0.124 | 7.237 ± 0.191 |
1.944 ± 0.095 | 5.062 ± 0.119 |
5.757 ± 0.19 | 7.668 ± 0.137 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.773 ± 0.096 | 4.054 ± 0.124 |
4.235 ± 0.106 | 3.449 ± 0.097 |
5.416 ± 0.137 | 6.167 ± 0.164 |
5.852 ± 0.172 | 7.437 ± 0.159 |
1.596 ± 0.083 | 3.478 ± 0.109 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
