
Raspberry bushy dwarf virus (isolate Malling Jewel raspberry/R15) (RBDV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Mayoviridae; Idaeovirus; Raspberry bushy dwarf virus
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
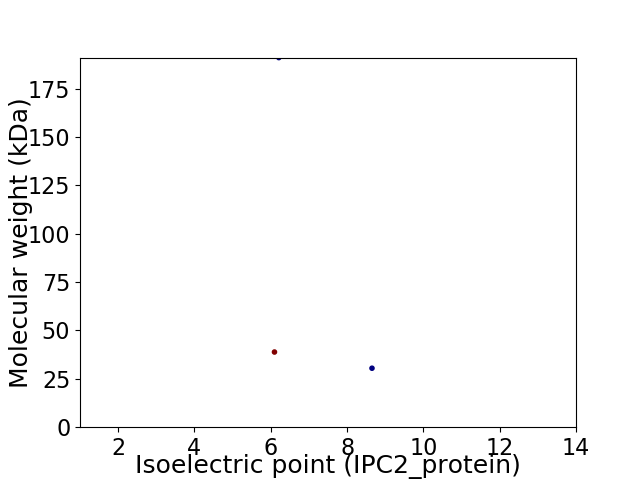
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q86564|CAPSD_RBDVR Capsid protein OS=Raspberry bushy dwarf virus (isolate Malling Jewel raspberry/R15) OX=652675 PE=4 SV=1
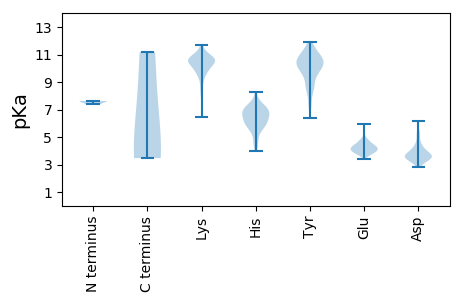
MM1 pKa = 7.62FSRR4 pKa = 11.84SSSTRR9 pKa = 11.84SSLVGSRR16 pKa = 11.84SGSIFGGGSVKK27 pKa = 10.39KK28 pKa = 10.33SSTVRR33 pKa = 11.84GFSAGLEE40 pKa = 4.08RR41 pKa = 11.84SRR43 pKa = 11.84GLPSASAGEE52 pKa = 4.19NQISLPGLRR61 pKa = 11.84IPVKK65 pKa = 10.64ASSQPGNYY73 pKa = 8.74YY74 pKa = 10.51LKK76 pKa = 10.55EE77 pKa = 4.12RR78 pKa = 11.84GIDD81 pKa = 3.63LPIVQQQKK89 pKa = 10.17FLAADD94 pKa = 3.7GKK96 pKa = 11.42EE97 pKa = 3.81MGEE100 pKa = 4.58CYY102 pKa = 10.83LLDD105 pKa = 3.61TSRR108 pKa = 11.84TDD110 pKa = 4.17LLDD113 pKa = 3.75AAKK116 pKa = 10.43AALNEE121 pKa = 4.4SNLLEE126 pKa = 4.07FNKK129 pKa = 10.11FKK131 pKa = 10.77EE132 pKa = 4.18FKK134 pKa = 10.22KK135 pKa = 11.08YY136 pKa = 9.93KK137 pKa = 9.95GKK139 pKa = 10.27NNEE142 pKa = 3.71FSLVEE147 pKa = 3.65ASVFDD152 pKa = 3.75KK153 pKa = 11.17LIRR156 pKa = 11.84KK157 pKa = 9.29DD158 pKa = 3.8DD159 pKa = 4.0SPIHH163 pKa = 6.27LNRR166 pKa = 11.84LLIAVLPAVGKK177 pKa = 7.54GTPGTARR184 pKa = 11.84IKK186 pKa = 10.63IRR188 pKa = 11.84DD189 pKa = 3.44ARR191 pKa = 11.84LDD193 pKa = 3.69DD194 pKa = 4.82GYY196 pKa = 11.89GEE198 pKa = 4.73LFSSEE203 pKa = 3.89NRR205 pKa = 11.84VDD207 pKa = 3.62SGYY210 pKa = 10.25IYY212 pKa = 9.88CINVGYY218 pKa = 9.87SVPKK222 pKa = 10.58SEE224 pKa = 4.7IDD226 pKa = 3.37YY227 pKa = 10.67KK228 pKa = 11.19INIDD232 pKa = 3.69FAGVPIKK239 pKa = 10.61DD240 pKa = 3.58GKK242 pKa = 10.25SPIWVKK248 pKa = 10.66AAFSLAGGPPVFLDD262 pKa = 3.54GTMSLGAEE270 pKa = 4.28ILPDD274 pKa = 3.39SHH276 pKa = 8.24KK277 pKa = 10.95EE278 pKa = 3.84LLGTSALLLNEE289 pKa = 4.37ANSNRR294 pKa = 11.84KK295 pKa = 9.19SFSGDD300 pKa = 3.2DD301 pKa = 3.49GEE303 pKa = 5.03LRR305 pKa = 11.84RR306 pKa = 11.84DD307 pKa = 3.5YY308 pKa = 10.42PYY310 pKa = 11.31KK311 pKa = 10.25RR312 pKa = 11.84FEE314 pKa = 4.79EE315 pKa = 4.26ISPLDD320 pKa = 4.19SISQVDD326 pKa = 3.81TASQDD331 pKa = 3.4SVNEE335 pKa = 3.97VNTEE339 pKa = 3.72NVQNGTGEE347 pKa = 4.46VYY349 pKa = 10.73LAPPSHH355 pKa = 6.52SVYY358 pKa = 11.18
MM1 pKa = 7.62FSRR4 pKa = 11.84SSSTRR9 pKa = 11.84SSLVGSRR16 pKa = 11.84SGSIFGGGSVKK27 pKa = 10.39KK28 pKa = 10.33SSTVRR33 pKa = 11.84GFSAGLEE40 pKa = 4.08RR41 pKa = 11.84SRR43 pKa = 11.84GLPSASAGEE52 pKa = 4.19NQISLPGLRR61 pKa = 11.84IPVKK65 pKa = 10.64ASSQPGNYY73 pKa = 8.74YY74 pKa = 10.51LKK76 pKa = 10.55EE77 pKa = 4.12RR78 pKa = 11.84GIDD81 pKa = 3.63LPIVQQQKK89 pKa = 10.17FLAADD94 pKa = 3.7GKK96 pKa = 11.42EE97 pKa = 3.81MGEE100 pKa = 4.58CYY102 pKa = 10.83LLDD105 pKa = 3.61TSRR108 pKa = 11.84TDD110 pKa = 4.17LLDD113 pKa = 3.75AAKK116 pKa = 10.43AALNEE121 pKa = 4.4SNLLEE126 pKa = 4.07FNKK129 pKa = 10.11FKK131 pKa = 10.77EE132 pKa = 4.18FKK134 pKa = 10.22KK135 pKa = 11.08YY136 pKa = 9.93KK137 pKa = 9.95GKK139 pKa = 10.27NNEE142 pKa = 3.71FSLVEE147 pKa = 3.65ASVFDD152 pKa = 3.75KK153 pKa = 11.17LIRR156 pKa = 11.84KK157 pKa = 9.29DD158 pKa = 3.8DD159 pKa = 4.0SPIHH163 pKa = 6.27LNRR166 pKa = 11.84LLIAVLPAVGKK177 pKa = 7.54GTPGTARR184 pKa = 11.84IKK186 pKa = 10.63IRR188 pKa = 11.84DD189 pKa = 3.44ARR191 pKa = 11.84LDD193 pKa = 3.69DD194 pKa = 4.82GYY196 pKa = 11.89GEE198 pKa = 4.73LFSSEE203 pKa = 3.89NRR205 pKa = 11.84VDD207 pKa = 3.62SGYY210 pKa = 10.25IYY212 pKa = 9.88CINVGYY218 pKa = 9.87SVPKK222 pKa = 10.58SEE224 pKa = 4.7IDD226 pKa = 3.37YY227 pKa = 10.67KK228 pKa = 11.19INIDD232 pKa = 3.69FAGVPIKK239 pKa = 10.61DD240 pKa = 3.58GKK242 pKa = 10.25SPIWVKK248 pKa = 10.66AAFSLAGGPPVFLDD262 pKa = 3.54GTMSLGAEE270 pKa = 4.28ILPDD274 pKa = 3.39SHH276 pKa = 8.24KK277 pKa = 10.95EE278 pKa = 3.84LLGTSALLLNEE289 pKa = 4.37ANSNRR294 pKa = 11.84KK295 pKa = 9.19SFSGDD300 pKa = 3.2DD301 pKa = 3.49GEE303 pKa = 5.03LRR305 pKa = 11.84RR306 pKa = 11.84DD307 pKa = 3.5YY308 pKa = 10.42PYY310 pKa = 11.31KK311 pKa = 10.25RR312 pKa = 11.84FEE314 pKa = 4.79EE315 pKa = 4.26ISPLDD320 pKa = 4.19SISQVDD326 pKa = 3.81TASQDD331 pKa = 3.4SVNEE335 pKa = 3.97VNTEE339 pKa = 3.72NVQNGTGEE347 pKa = 4.46VYY349 pKa = 10.73LAPPSHH355 pKa = 6.52SVYY358 pKa = 11.18
Molecular weight: 38.87 kDa
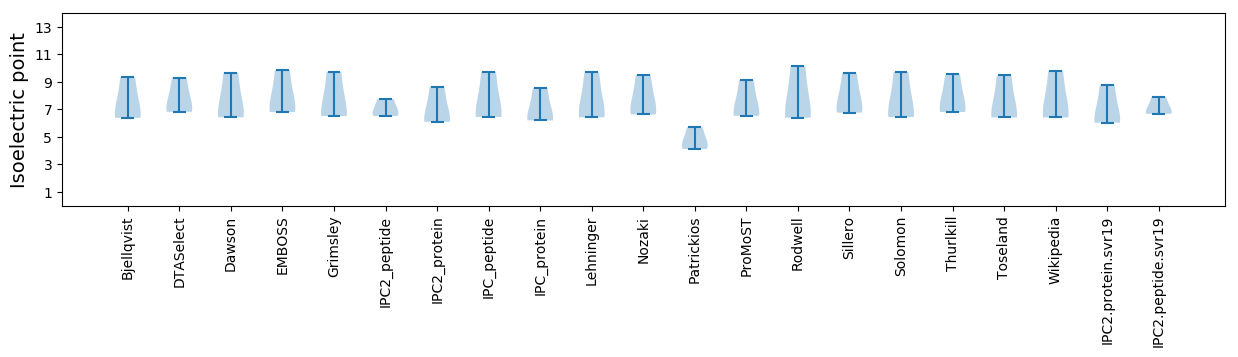
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q86564|CAPSD_RBDVR Capsid protein OS=Raspberry bushy dwarf virus (isolate Malling Jewel raspberry/R15) OX=652675 PE=4 SV=1
MM1 pKa = 7.4SKK3 pKa = 10.45KK4 pKa = 10.15AVPPIVKK11 pKa = 9.74AQYY14 pKa = 9.74EE15 pKa = 4.47LYY17 pKa = 10.33NRR19 pKa = 11.84KK20 pKa = 9.49LNRR23 pKa = 11.84AIKK26 pKa = 10.6VSGSQKK32 pKa = 10.72KK33 pKa = 10.18LDD35 pKa = 3.55ASFVGFSEE43 pKa = 4.29SSNPEE48 pKa = 3.6TGKK51 pKa = 9.35PHH53 pKa = 7.55ADD55 pKa = 2.83MSMSAKK61 pKa = 9.62VKK63 pKa = 10.4RR64 pKa = 11.84VNTWLKK70 pKa = 10.92NFDD73 pKa = 3.34RR74 pKa = 11.84EE75 pKa = 4.07YY76 pKa = 10.32WDD78 pKa = 3.32NQFASKK84 pKa = 10.1PIPRR88 pKa = 11.84PAKK91 pKa = 9.57QVLKK95 pKa = 10.49GSSSKK100 pKa = 10.55SQQRR104 pKa = 11.84DD105 pKa = 2.99EE106 pKa = 4.84GEE108 pKa = 4.29VVFTRR113 pKa = 11.84KK114 pKa = 9.98DD115 pKa = 3.33SQKK118 pKa = 10.42SVRR121 pKa = 11.84TVSYY125 pKa = 8.48WVCTPEE131 pKa = 5.89KK132 pKa = 10.67SMKK135 pKa = 8.3PLKK138 pKa = 10.58YY139 pKa = 10.24KK140 pKa = 10.2EE141 pKa = 4.42DD142 pKa = 3.79EE143 pKa = 4.33NVVEE147 pKa = 4.45VTFNDD152 pKa = 3.91LAAQKK157 pKa = 10.75AGDD160 pKa = 3.89KK161 pKa = 10.47LVSILLEE168 pKa = 4.07INVVGGAVDD177 pKa = 3.46DD178 pKa = 4.31KK179 pKa = 11.67GRR181 pKa = 11.84VAVLEE186 pKa = 3.9KK187 pKa = 10.72DD188 pKa = 3.18AAVTVDD194 pKa = 3.79YY195 pKa = 11.19LLGSPYY201 pKa = 10.51EE202 pKa = 4.47AINLVSGLNKK212 pKa = 9.97INFRR216 pKa = 11.84SMTDD220 pKa = 3.24VVDD223 pKa = 5.42SIPSLLNEE231 pKa = 3.95RR232 pKa = 11.84KK233 pKa = 9.88VCVFQNDD240 pKa = 3.94DD241 pKa = 3.37SSSFYY246 pKa = 9.67IRR248 pKa = 11.84KK249 pKa = 7.9WANFLQEE256 pKa = 3.96VSAVLPVGTGKK267 pKa = 10.68SSTIVLTT274 pKa = 3.95
MM1 pKa = 7.4SKK3 pKa = 10.45KK4 pKa = 10.15AVPPIVKK11 pKa = 9.74AQYY14 pKa = 9.74EE15 pKa = 4.47LYY17 pKa = 10.33NRR19 pKa = 11.84KK20 pKa = 9.49LNRR23 pKa = 11.84AIKK26 pKa = 10.6VSGSQKK32 pKa = 10.72KK33 pKa = 10.18LDD35 pKa = 3.55ASFVGFSEE43 pKa = 4.29SSNPEE48 pKa = 3.6TGKK51 pKa = 9.35PHH53 pKa = 7.55ADD55 pKa = 2.83MSMSAKK61 pKa = 9.62VKK63 pKa = 10.4RR64 pKa = 11.84VNTWLKK70 pKa = 10.92NFDD73 pKa = 3.34RR74 pKa = 11.84EE75 pKa = 4.07YY76 pKa = 10.32WDD78 pKa = 3.32NQFASKK84 pKa = 10.1PIPRR88 pKa = 11.84PAKK91 pKa = 9.57QVLKK95 pKa = 10.49GSSSKK100 pKa = 10.55SQQRR104 pKa = 11.84DD105 pKa = 2.99EE106 pKa = 4.84GEE108 pKa = 4.29VVFTRR113 pKa = 11.84KK114 pKa = 9.98DD115 pKa = 3.33SQKK118 pKa = 10.42SVRR121 pKa = 11.84TVSYY125 pKa = 8.48WVCTPEE131 pKa = 5.89KK132 pKa = 10.67SMKK135 pKa = 8.3PLKK138 pKa = 10.58YY139 pKa = 10.24KK140 pKa = 10.2EE141 pKa = 4.42DD142 pKa = 3.79EE143 pKa = 4.33NVVEE147 pKa = 4.45VTFNDD152 pKa = 3.91LAAQKK157 pKa = 10.75AGDD160 pKa = 3.89KK161 pKa = 10.47LVSILLEE168 pKa = 4.07INVVGGAVDD177 pKa = 3.46DD178 pKa = 4.31KK179 pKa = 11.67GRR181 pKa = 11.84VAVLEE186 pKa = 3.9KK187 pKa = 10.72DD188 pKa = 3.18AAVTVDD194 pKa = 3.79YY195 pKa = 11.19LLGSPYY201 pKa = 10.51EE202 pKa = 4.47AINLVSGLNKK212 pKa = 9.97INFRR216 pKa = 11.84SMTDD220 pKa = 3.24VVDD223 pKa = 5.42SIPSLLNEE231 pKa = 3.95RR232 pKa = 11.84KK233 pKa = 9.88VCVFQNDD240 pKa = 3.94DD241 pKa = 3.37SSSFYY246 pKa = 9.67IRR248 pKa = 11.84KK249 pKa = 7.9WANFLQEE256 pKa = 3.96VSAVLPVGTGKK267 pKa = 10.68SSTIVLTT274 pKa = 3.95
Molecular weight: 30.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2326 |
274 |
1694 |
775.3 |
86.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.89 ± 0.359 | 1.978 ± 0.639 |
6.793 ± 0.224 | 5.374 ± 0.244 |
4.772 ± 0.4 | 6.105 ± 0.835 |
2.107 ± 0.708 | 5.546 ± 0.484 |
6.922 ± 0.954 | 9.114 ± 0.418 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.677 ± 0.236 | 4.729 ± 0.246 |
4.385 ± 0.192 | 2.537 ± 0.259 |
5.245 ± 0.216 | 9.329 ± 1.139 |
5.116 ± 0.783 | 7.696 ± 0.921 |
1.075 ± 0.213 | 3.611 ± 0.176 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
