
Bat polyomavirus 5a
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus; Dobsonia moluccensis polyomavirus 1
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
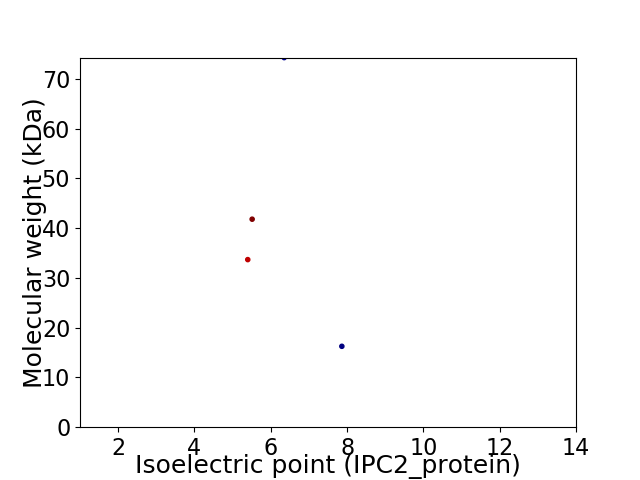
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
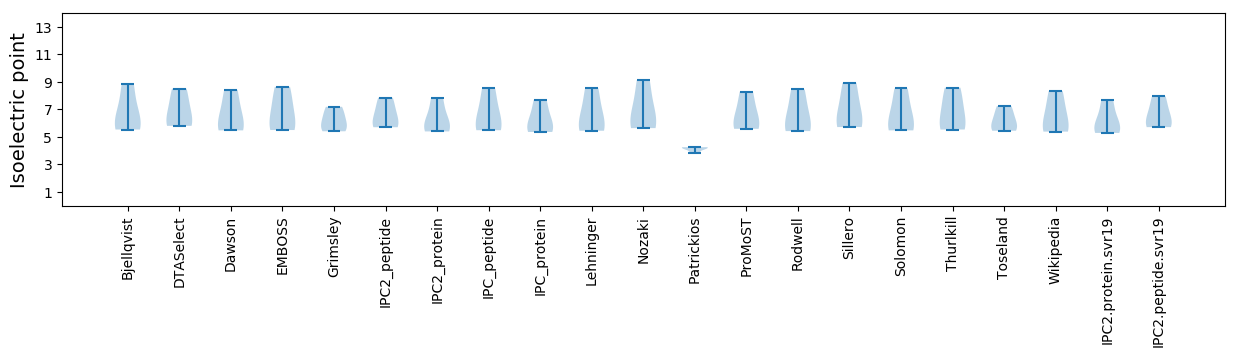
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D5ZZ35|A0A0D5ZZ35_9POLY Capsid protein VP1 OS=Bat polyomavirus 5a OX=1623687 GN=VP1 PE=3 SV=1
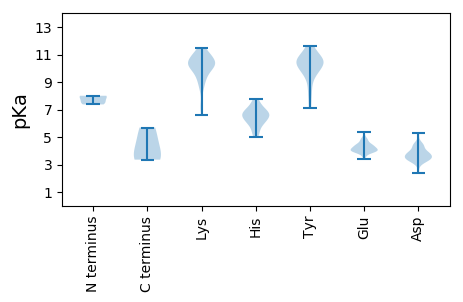
MM1 pKa = 7.37APKK4 pKa = 10.38RR5 pKa = 11.84KK6 pKa = 9.72GGSSKK11 pKa = 11.35GEE13 pKa = 3.93TKK15 pKa = 10.4SCSNICSTKK24 pKa = 10.12CPVVNPVPKK33 pKa = 10.43LLVKK37 pKa = 10.82GGIQVLSVKK46 pKa = 9.56TGPDD50 pKa = 3.54SITQIEE56 pKa = 4.95AYY58 pKa = 10.06LNPRR62 pKa = 11.84MGQPPASSTLGPGYY76 pKa = 9.07SEE78 pKa = 4.25KK79 pKa = 11.14VGVATYY85 pKa = 9.61WGDD88 pKa = 3.39TDD90 pKa = 5.09KK91 pKa = 11.24PLANHH96 pKa = 6.99LPMYY100 pKa = 10.02SCARR104 pKa = 11.84INLPMLNEE112 pKa = 4.9DD113 pKa = 3.86MTCDD117 pKa = 3.43TLQMWEE123 pKa = 4.04AVSVKK128 pKa = 9.81TEE130 pKa = 4.11VVGTTSLLDD139 pKa = 3.1SHH141 pKa = 6.69YY142 pKa = 10.65AGKK145 pKa = 10.55RR146 pKa = 11.84MYY148 pKa = 10.51SDD150 pKa = 3.44YY151 pKa = 11.34GIALPIEE158 pKa = 4.67GTSLHH163 pKa = 6.1FFSVGGEE170 pKa = 3.95PLDD173 pKa = 4.2LQGLVANGKK182 pKa = 9.84AIYY185 pKa = 7.17PTTLAVINNMTEE197 pKa = 4.06KK198 pKa = 10.89NQVLQQKK205 pKa = 10.33AKK207 pKa = 10.98AKK209 pKa = 9.68LLKK212 pKa = 10.76DD213 pKa = 3.07GTYY216 pKa = 9.74PVEE219 pKa = 3.95MWGPDD224 pKa = 3.26PSRR227 pKa = 11.84NEE229 pKa = 3.41NTRR232 pKa = 11.84YY233 pKa = 10.37YY234 pKa = 10.51GTFTGGQTTPPVLQFTNTVTTVLLDD259 pKa = 3.84EE260 pKa = 5.12NGVGPLCKK268 pKa = 10.13GDD270 pKa = 3.88GLFIASVDD278 pKa = 3.81VIGFYY283 pKa = 10.37TDD285 pKa = 2.56SSGYY289 pKa = 8.56MYY291 pKa = 10.98YY292 pKa = 10.34RR293 pKa = 11.84GLPRR297 pKa = 11.84YY298 pKa = 9.74FNITLRR304 pKa = 11.84KK305 pKa = 9.42RR306 pKa = 11.84NVKK309 pKa = 10.03NPYY312 pKa = 9.3PVTSLLSSLFNNMIPKK328 pKa = 9.91LQGQPMEE335 pKa = 4.42GEE337 pKa = 4.05EE338 pKa = 4.43GQVEE342 pKa = 4.38EE343 pKa = 4.18VRR345 pKa = 11.84VYY347 pKa = 10.83EE348 pKa = 4.26GLEE351 pKa = 3.96GLPGDD356 pKa = 4.44PDD358 pKa = 2.97MDD360 pKa = 3.57RR361 pKa = 11.84YY362 pKa = 10.82VDD364 pKa = 4.43QFGQDD369 pKa = 3.12KK370 pKa = 10.25TIIPSLRR377 pKa = 11.84TNDD380 pKa = 3.51EE381 pKa = 4.01LL382 pKa = 5.69
MM1 pKa = 7.37APKK4 pKa = 10.38RR5 pKa = 11.84KK6 pKa = 9.72GGSSKK11 pKa = 11.35GEE13 pKa = 3.93TKK15 pKa = 10.4SCSNICSTKK24 pKa = 10.12CPVVNPVPKK33 pKa = 10.43LLVKK37 pKa = 10.82GGIQVLSVKK46 pKa = 9.56TGPDD50 pKa = 3.54SITQIEE56 pKa = 4.95AYY58 pKa = 10.06LNPRR62 pKa = 11.84MGQPPASSTLGPGYY76 pKa = 9.07SEE78 pKa = 4.25KK79 pKa = 11.14VGVATYY85 pKa = 9.61WGDD88 pKa = 3.39TDD90 pKa = 5.09KK91 pKa = 11.24PLANHH96 pKa = 6.99LPMYY100 pKa = 10.02SCARR104 pKa = 11.84INLPMLNEE112 pKa = 4.9DD113 pKa = 3.86MTCDD117 pKa = 3.43TLQMWEE123 pKa = 4.04AVSVKK128 pKa = 9.81TEE130 pKa = 4.11VVGTTSLLDD139 pKa = 3.1SHH141 pKa = 6.69YY142 pKa = 10.65AGKK145 pKa = 10.55RR146 pKa = 11.84MYY148 pKa = 10.51SDD150 pKa = 3.44YY151 pKa = 11.34GIALPIEE158 pKa = 4.67GTSLHH163 pKa = 6.1FFSVGGEE170 pKa = 3.95PLDD173 pKa = 4.2LQGLVANGKK182 pKa = 9.84AIYY185 pKa = 7.17PTTLAVINNMTEE197 pKa = 4.06KK198 pKa = 10.89NQVLQQKK205 pKa = 10.33AKK207 pKa = 10.98AKK209 pKa = 9.68LLKK212 pKa = 10.76DD213 pKa = 3.07GTYY216 pKa = 9.74PVEE219 pKa = 3.95MWGPDD224 pKa = 3.26PSRR227 pKa = 11.84NEE229 pKa = 3.41NTRR232 pKa = 11.84YY233 pKa = 10.37YY234 pKa = 10.51GTFTGGQTTPPVLQFTNTVTTVLLDD259 pKa = 3.84EE260 pKa = 5.12NGVGPLCKK268 pKa = 10.13GDD270 pKa = 3.88GLFIASVDD278 pKa = 3.81VIGFYY283 pKa = 10.37TDD285 pKa = 2.56SSGYY289 pKa = 8.56MYY291 pKa = 10.98YY292 pKa = 10.34RR293 pKa = 11.84GLPRR297 pKa = 11.84YY298 pKa = 9.74FNITLRR304 pKa = 11.84KK305 pKa = 9.42RR306 pKa = 11.84NVKK309 pKa = 10.03NPYY312 pKa = 9.3PVTSLLSSLFNNMIPKK328 pKa = 9.91LQGQPMEE335 pKa = 4.42GEE337 pKa = 4.05EE338 pKa = 4.43GQVEE342 pKa = 4.38EE343 pKa = 4.18VRR345 pKa = 11.84VYY347 pKa = 10.83EE348 pKa = 4.26GLEE351 pKa = 3.96GLPGDD356 pKa = 4.44PDD358 pKa = 2.97MDD360 pKa = 3.57RR361 pKa = 11.84YY362 pKa = 10.82VDD364 pKa = 4.43QFGQDD369 pKa = 3.12KK370 pKa = 10.25TIIPSLRR377 pKa = 11.84TNDD380 pKa = 3.51EE381 pKa = 4.01LL382 pKa = 5.69
Molecular weight: 41.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D5ZYM7|A0A0D5ZYM7_9POLY Large T antigen OS=Bat polyomavirus 5a OX=1623687 PE=4 SV=1
MM1 pKa = 8.0DD2 pKa = 4.39KK3 pKa = 11.03FMDD6 pKa = 3.86RR7 pKa = 11.84EE8 pKa = 4.06EE9 pKa = 4.49LKK11 pKa = 10.7EE12 pKa = 3.99LCEE15 pKa = 4.09LLNIPAHH22 pKa = 6.64CYY24 pKa = 10.85GNLPMMKK31 pKa = 9.53INYY34 pKa = 9.78KK35 pKa = 10.14KK36 pKa = 9.33MCLIYY41 pKa = 10.7HH42 pKa = 6.89PDD44 pKa = 3.28KK45 pKa = 11.45GGDD48 pKa = 3.42VAKK51 pKa = 8.75MQRR54 pKa = 11.84MNEE57 pKa = 3.86LWQKK61 pKa = 10.39LQDD64 pKa = 3.73GVINARR70 pKa = 11.84DD71 pKa = 3.8EE72 pKa = 4.8GPVSRR77 pKa = 11.84WFWEE81 pKa = 4.08YY82 pKa = 10.63QGQTLRR88 pKa = 11.84EE89 pKa = 3.87FLGPDD94 pKa = 3.21FNKK97 pKa = 10.11RR98 pKa = 11.84FCKK101 pKa = 10.34VFPTCLYY108 pKa = 10.53ASKK111 pKa = 10.27EE112 pKa = 3.93FCFCVCCLLNKK123 pKa = 7.32QHH125 pKa = 7.21KK126 pKa = 8.13IYY128 pKa = 10.29KK129 pKa = 9.17VKK131 pKa = 10.59RR132 pKa = 11.84EE133 pKa = 3.92KK134 pKa = 10.81NAA136 pKa = 3.36
MM1 pKa = 8.0DD2 pKa = 4.39KK3 pKa = 11.03FMDD6 pKa = 3.86RR7 pKa = 11.84EE8 pKa = 4.06EE9 pKa = 4.49LKK11 pKa = 10.7EE12 pKa = 3.99LCEE15 pKa = 4.09LLNIPAHH22 pKa = 6.64CYY24 pKa = 10.85GNLPMMKK31 pKa = 9.53INYY34 pKa = 9.78KK35 pKa = 10.14KK36 pKa = 9.33MCLIYY41 pKa = 10.7HH42 pKa = 6.89PDD44 pKa = 3.28KK45 pKa = 11.45GGDD48 pKa = 3.42VAKK51 pKa = 8.75MQRR54 pKa = 11.84MNEE57 pKa = 3.86LWQKK61 pKa = 10.39LQDD64 pKa = 3.73GVINARR70 pKa = 11.84DD71 pKa = 3.8EE72 pKa = 4.8GPVSRR77 pKa = 11.84WFWEE81 pKa = 4.08YY82 pKa = 10.63QGQTLRR88 pKa = 11.84EE89 pKa = 3.87FLGPDD94 pKa = 3.21FNKK97 pKa = 10.11RR98 pKa = 11.84FCKK101 pKa = 10.34VFPTCLYY108 pKa = 10.53ASKK111 pKa = 10.27EE112 pKa = 3.93FCFCVCCLLNKK123 pKa = 7.32QHH125 pKa = 7.21KK126 pKa = 8.13IYY128 pKa = 10.29KK129 pKa = 9.17VKK131 pKa = 10.59RR132 pKa = 11.84EE133 pKa = 3.92KK134 pKa = 10.81NAA136 pKa = 3.36
Molecular weight: 16.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1474 |
136 |
647 |
368.5 |
41.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.834 ± 1.534 | 2.307 ± 0.752 |
5.02 ± 0.174 | 6.716 ± 0.509 |
4.071 ± 0.695 | 6.513 ± 1.16 |
1.628 ± 0.261 | 4.953 ± 0.517 |
7.123 ± 1.216 | 10.176 ± 0.419 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.917 ± 0.508 | 5.563 ± 0.656 |
5.834 ± 0.528 | 4.478 ± 0.203 |
4.206 ± 0.472 | 5.97 ± 0.756 |
5.563 ± 0.815 | 6.174 ± 0.527 |
1.153 ± 0.324 | 3.799 ± 0.453 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
