
Hubei macula-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
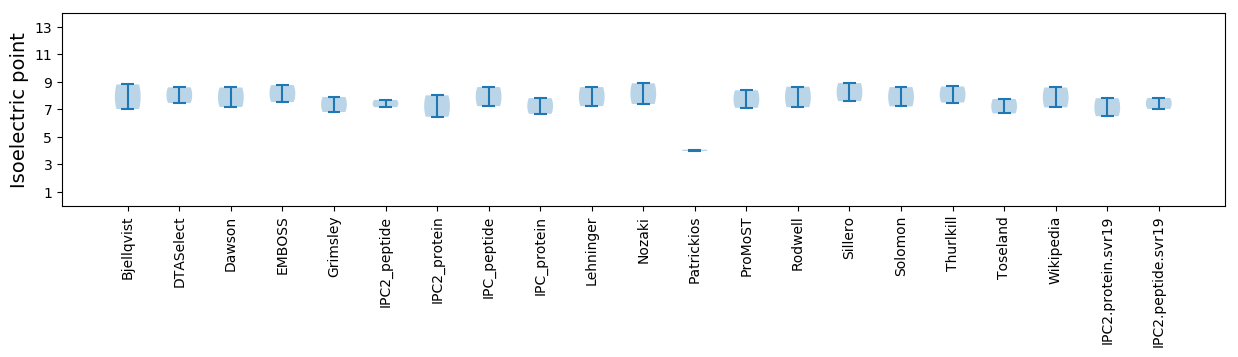
Average proteome isoelectric point is 7.16
Get precalculated fractions of proteins
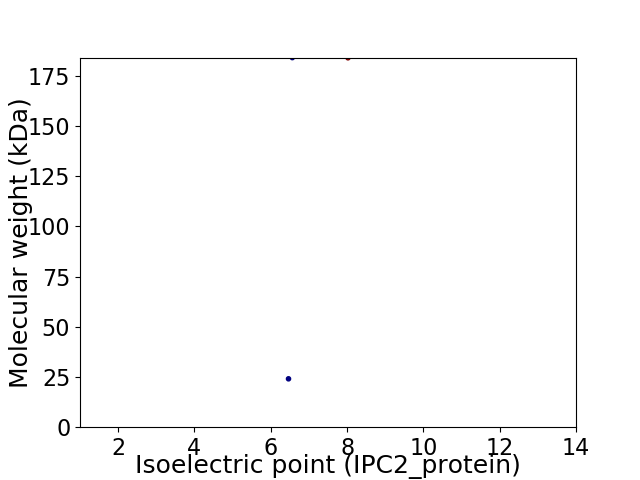
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJV4|A0A1L3KJV4_9VIRU Uncharacterized protein OS=Hubei macula-like virus 2 OX=1922923 PE=4 SV=1
MM1 pKa = 6.94NTLLQTAIEE10 pKa = 4.21HH11 pKa = 6.22APAIISALPSLLPPSSQSTSTSSDD35 pKa = 3.26PVHH38 pKa = 7.91DD39 pKa = 5.4DD40 pKa = 3.47RR41 pKa = 11.84LLLANSALPPAPSTPSHH58 pKa = 6.38ISSFPSSSSVKK69 pKa = 10.46VPFQYY74 pKa = 10.99VWFGLDD80 pKa = 3.15GTEE83 pKa = 4.65SKK85 pKa = 10.5NASITIPEE93 pKa = 4.1IPSVKK98 pKa = 10.18KK99 pKa = 10.34IIAFFRR105 pKa = 11.84DD106 pKa = 3.33AKK108 pKa = 10.63LVNLEE113 pKa = 3.97AAVYY117 pKa = 8.67PHH119 pKa = 7.03FDD121 pKa = 3.85CLEE124 pKa = 4.01KK125 pKa = 10.62PITVDD130 pKa = 4.84LMWSPSDD137 pKa = 3.68TVPSGSILNCPGAVCFTVGGQFLSQSGILPCDD169 pKa = 3.84LNFMNPIIKK178 pKa = 10.41SPIPYY183 pKa = 8.88TNHH186 pKa = 6.0PRR188 pKa = 11.84LNAKK192 pKa = 9.7FYY194 pKa = 11.17KK195 pKa = 10.4NQTEE199 pKa = 4.33LQSHH203 pKa = 6.51RR204 pKa = 11.84KK205 pKa = 7.15ATIIIRR211 pKa = 11.84GIIACAHH218 pKa = 5.19PTVYY222 pKa = 10.85AA223 pKa = 4.13
MM1 pKa = 6.94NTLLQTAIEE10 pKa = 4.21HH11 pKa = 6.22APAIISALPSLLPPSSQSTSTSSDD35 pKa = 3.26PVHH38 pKa = 7.91DD39 pKa = 5.4DD40 pKa = 3.47RR41 pKa = 11.84LLLANSALPPAPSTPSHH58 pKa = 6.38ISSFPSSSSVKK69 pKa = 10.46VPFQYY74 pKa = 10.99VWFGLDD80 pKa = 3.15GTEE83 pKa = 4.65SKK85 pKa = 10.5NASITIPEE93 pKa = 4.1IPSVKK98 pKa = 10.18KK99 pKa = 10.34IIAFFRR105 pKa = 11.84DD106 pKa = 3.33AKK108 pKa = 10.63LVNLEE113 pKa = 3.97AAVYY117 pKa = 8.67PHH119 pKa = 7.03FDD121 pKa = 3.85CLEE124 pKa = 4.01KK125 pKa = 10.62PITVDD130 pKa = 4.84LMWSPSDD137 pKa = 3.68TVPSGSILNCPGAVCFTVGGQFLSQSGILPCDD169 pKa = 3.84LNFMNPIIKK178 pKa = 10.41SPIPYY183 pKa = 8.88TNHH186 pKa = 6.0PRR188 pKa = 11.84LNAKK192 pKa = 9.7FYY194 pKa = 11.17KK195 pKa = 10.4NQTEE199 pKa = 4.33LQSHH203 pKa = 6.51RR204 pKa = 11.84KK205 pKa = 7.15ATIIIRR211 pKa = 11.84GIIACAHH218 pKa = 5.19PTVYY222 pKa = 10.85AA223 pKa = 4.13
Molecular weight: 24.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJV4|A0A1L3KJV4_9VIRU Uncharacterized protein OS=Hubei macula-like virus 2 OX=1922923 PE=4 SV=1
MM1 pKa = 7.65SSLSNNMQILSNTLHH16 pKa = 7.02KK17 pKa = 10.71DD18 pKa = 3.19ASTHH22 pKa = 6.19PLLTHH27 pKa = 5.67TLKK30 pKa = 10.8NLHH33 pKa = 6.26TSLHH37 pKa = 5.81AFPWSLTSEE46 pKa = 4.21QVEE49 pKa = 4.15LLVNLGINVSPHH61 pKa = 4.72GTLPHH66 pKa = 6.16PHH68 pKa = 7.12PVHH71 pKa = 6.5KK72 pKa = 10.63CIEE75 pKa = 4.12THH77 pKa = 6.21VLFNHH82 pKa = 6.3WNHH85 pKa = 6.76LAVQPSAVCFMKK97 pKa = 10.38PSKK100 pKa = 9.85FAKK103 pKa = 10.19LSNSNPNFIEE113 pKa = 3.78LHH115 pKa = 6.05NYY117 pKa = 9.43IIDD120 pKa = 3.7ARR122 pKa = 11.84DD123 pKa = 3.39PTRR126 pKa = 11.84YY127 pKa = 9.35PSISSSLPTTPYY139 pKa = 11.17LFIHH143 pKa = 7.15DD144 pKa = 4.48ALMFISPNEE153 pKa = 4.19IIDD156 pKa = 4.46LFSNCPNLTHH166 pKa = 6.97VFGTLVLPAEE176 pKa = 4.94SIHH179 pKa = 6.42NIPSFYY185 pKa = 10.07PDD187 pKa = 2.93IYY189 pKa = 10.43TYY191 pKa = 10.71HH192 pKa = 6.89HH193 pKa = 6.94EE194 pKa = 4.5NGNLHH199 pKa = 6.39YY200 pKa = 9.97FLEE203 pKa = 5.26GNPSPHH209 pKa = 5.19YY210 pKa = 7.7TQPKK214 pKa = 9.34DD215 pKa = 3.14AAIWLTTHH223 pKa = 7.56TINGPNFSLSVVILEE238 pKa = 4.39SWLSVHH244 pKa = 7.05SILITKK250 pKa = 7.11YY251 pKa = 8.0QPRR254 pKa = 11.84VPTYY258 pKa = 8.99KK259 pKa = 10.25HH260 pKa = 5.71YY261 pKa = 10.78FRR263 pKa = 11.84IPKK266 pKa = 9.42TSLLPNPTNLNLPLHH281 pKa = 6.16SRR283 pKa = 11.84LVPTEE288 pKa = 3.57VHH290 pKa = 6.58ASLFNYY296 pKa = 8.49VRR298 pKa = 11.84AVRR301 pKa = 11.84TLRR304 pKa = 11.84TSDD307 pKa = 3.08PSAFIRR313 pKa = 11.84SLRR316 pKa = 11.84SKK318 pKa = 10.86EE319 pKa = 3.8EE320 pKa = 3.84YY321 pKa = 9.87RR322 pKa = 11.84WVHH325 pKa = 5.97ASAWDD330 pKa = 3.6QLVQFALATHH340 pKa = 6.44NVTSPSLFAFIEE352 pKa = 4.27TFLRR356 pKa = 11.84QILNHH361 pKa = 6.63AKK363 pKa = 9.04LTFLQFIPNPIHH375 pKa = 7.1HH376 pKa = 6.91AFFWLLAKK384 pKa = 10.16ALSYY388 pKa = 8.97ITPKK392 pKa = 10.04PYY394 pKa = 9.56STIFHH399 pKa = 5.94VASIISPLLHH409 pKa = 6.29TYY411 pKa = 8.99WLFSHH416 pKa = 7.45PSPPQVILANYY427 pKa = 7.89NQYY430 pKa = 10.45FHH432 pKa = 7.83PDD434 pKa = 3.0PWLLFADD441 pKa = 4.96RR442 pKa = 11.84YY443 pKa = 10.85SIVAMPKK450 pKa = 9.66PFLSLTLQTPPPSPTSTPPPQPTQQTQPALPAPPPVQTSNPLNSTPFAPPSSLPPLQTPQSAQPTILSTSTPVLPPIVSPPITTCNNPDD539 pKa = 4.14PLLTDD544 pKa = 3.8PTAIGVILPFNQLYY558 pKa = 9.79PGLSMPSHH566 pKa = 6.2VCNFPSRR573 pKa = 11.84LRR575 pKa = 11.84DD576 pKa = 3.44PNAPLPSYY584 pKa = 10.48PITQVCLFDD593 pKa = 4.64ALSTLLQQPQHH604 pKa = 6.19NLFQILAQYY613 pKa = 10.61LPDD616 pKa = 3.82SLVVGPEE623 pKa = 4.05EE624 pKa = 4.18KK625 pKa = 10.11TSGYY629 pKa = 8.66STDD632 pKa = 3.74HH633 pKa = 7.16LDD635 pKa = 2.94ILLYY639 pKa = 10.9HH640 pKa = 6.95FDD642 pKa = 3.55QPLIIHH648 pKa = 6.62SSLGDD653 pKa = 3.62EE654 pKa = 3.92PHH656 pKa = 6.6GPKK659 pKa = 10.16NSRR662 pKa = 11.84NLLHH666 pKa = 7.15LYY668 pKa = 10.03HH669 pKa = 6.35FTGHH673 pKa = 6.66WSAQPPSQPPLRR685 pKa = 11.84GSGLSPFANAVLSFRR700 pKa = 11.84CEE702 pKa = 4.2DD703 pKa = 3.46NSLLPIANVYY713 pKa = 6.69VHH715 pKa = 6.49KK716 pKa = 10.99VNYY719 pKa = 9.36HH720 pKa = 4.97RR721 pKa = 11.84AKK723 pKa = 10.65NLSSNLKK730 pKa = 10.1NEE732 pKa = 3.93NDD734 pKa = 4.8GILLTQLKK742 pKa = 10.34QSVNDD747 pKa = 3.51NKK749 pKa = 10.76LLSKK753 pKa = 10.2LDD755 pKa = 3.64SMMDD759 pKa = 3.54NPVSRR764 pKa = 11.84HH765 pKa = 4.74VEE767 pKa = 4.32LIHH770 pKa = 6.66IAGFPGCGKK779 pKa = 9.18SHH781 pKa = 7.09PVASLLKK788 pKa = 10.27CKK790 pKa = 7.84TFRR793 pKa = 11.84NSFRR797 pKa = 11.84VVVPTVEE804 pKa = 5.63LRR806 pKa = 11.84SEE808 pKa = 4.07WKK810 pKa = 10.42DD811 pKa = 3.36KK812 pKa = 11.72LEE814 pKa = 4.62LPDD817 pKa = 3.83FEE819 pKa = 4.53SWRR822 pKa = 11.84ISTWEE827 pKa = 3.79TSLLKK832 pKa = 10.26LAKK835 pKa = 10.26VLVIDD840 pKa = 4.86EE841 pKa = 4.58VYY843 pKa = 10.82KK844 pKa = 11.03LPNGFLDD851 pKa = 4.12LAVALDD857 pKa = 3.71PTLQFVILLGDD868 pKa = 4.14PCQGTYY874 pKa = 10.6HH875 pKa = 6.6SLSPHH880 pKa = 5.2STNHH884 pKa = 6.13HH885 pKa = 6.83IYY887 pKa = 10.41PEE889 pKa = 4.03TEE891 pKa = 4.0YY892 pKa = 10.97LSKK895 pKa = 11.18YY896 pKa = 9.93RR897 pKa = 11.84DD898 pKa = 4.6FYY900 pKa = 11.32CLWTHH905 pKa = 6.54RR906 pKa = 11.84TPQVIANLLNVRR918 pKa = 11.84SSSKK922 pKa = 10.89EE923 pKa = 3.35NGQISRR929 pKa = 11.84SNTLNASFPVLATSMAIATHH949 pKa = 6.68LNQNGFRR956 pKa = 11.84SITYY960 pKa = 9.9AGSQGLTLNRR970 pKa = 11.84LTYY973 pKa = 10.63LCVDD977 pKa = 4.27RR978 pKa = 11.84YY979 pKa = 9.9ISLMSPGVTLVALTRR994 pKa = 11.84STKK997 pKa = 10.82GIIFNGNYY1005 pKa = 10.27DD1006 pKa = 3.84LLGNLGYY1013 pKa = 9.53TNTLLEE1019 pKa = 4.54SIFRR1023 pKa = 11.84GEE1025 pKa = 4.67KK1026 pKa = 9.33IDD1028 pKa = 3.9FQHH1031 pKa = 7.23LFHH1034 pKa = 6.12QQLRR1038 pKa = 11.84GSRR1041 pKa = 11.84IITQPIMSPTDD1052 pKa = 2.99RR1053 pKa = 11.84KK1054 pKa = 9.22MLRR1057 pKa = 11.84GGAILITKK1065 pKa = 6.86TAKK1068 pKa = 9.81IDD1070 pKa = 3.49INSSIDD1076 pKa = 3.55FLRR1079 pKa = 11.84QDD1081 pKa = 3.0SQLRR1085 pKa = 11.84SNPEE1089 pKa = 4.63LIPLHH1094 pKa = 7.06DD1095 pKa = 4.56NFYY1098 pKa = 10.5LPPSRR1103 pKa = 11.84LVLSQSFSSSLHH1115 pKa = 6.27PPAKK1119 pKa = 10.17PNNTPFAPTPFEE1131 pKa = 4.02PSYY1134 pKa = 10.37PGYY1137 pKa = 10.0PFEE1140 pKa = 4.92TYY1142 pKa = 9.85FANLPDD1148 pKa = 4.23PSEE1151 pKa = 4.5PEE1153 pKa = 4.52SKK1155 pKa = 10.56EE1156 pKa = 3.33KK1157 pKa = 9.74WYY1159 pKa = 10.59KK1160 pKa = 10.57DD1161 pKa = 3.0KK1162 pKa = 10.99RR1163 pKa = 11.84SQQFPILNKK1172 pKa = 9.91GSPYY1176 pKa = 10.87SPFPANIVAPIHH1188 pKa = 6.85DD1189 pKa = 4.4SKK1191 pKa = 11.46KK1192 pKa = 10.94DD1193 pKa = 3.57STLLPLSINKK1203 pKa = 9.63RR1204 pKa = 11.84LRR1206 pKa = 11.84FRR1208 pKa = 11.84PDD1210 pKa = 2.81NKK1212 pKa = 9.99PYY1214 pKa = 10.59QITEE1218 pKa = 3.89LDD1220 pKa = 3.72HH1221 pKa = 6.72YY1222 pKa = 10.01VANHH1226 pKa = 6.85LFQSYY1231 pKa = 10.48CEE1233 pKa = 4.19TLNTTIDD1240 pKa = 3.11IKK1242 pKa = 11.45VPFDD1246 pKa = 3.07AHH1248 pKa = 7.11LFAEE1252 pKa = 5.28CINLNDD1258 pKa = 3.76YY1259 pKa = 10.56NQLTKK1264 pKa = 9.61KK1265 pKa = 8.78TRR1267 pKa = 11.84ATLVANAYY1275 pKa = 10.2RR1276 pKa = 11.84SDD1278 pKa = 3.94PDD1280 pKa = 3.0WRR1282 pKa = 11.84FTFVRR1287 pKa = 11.84IFSKK1291 pKa = 9.02TQHH1294 pKa = 6.65KK1295 pKa = 10.69INTNSLYY1302 pKa = 10.29TSWKK1306 pKa = 9.38ACQTLALMNDD1316 pKa = 4.87LIVLTLGPVKK1326 pKa = 10.46KK1327 pKa = 10.05YY1328 pKa = 10.43QRR1330 pKa = 11.84LINDD1334 pKa = 4.47PLRR1337 pKa = 11.84KK1338 pKa = 9.92SNIFTYY1344 pKa = 10.39GGKK1347 pKa = 8.06TPFDD1351 pKa = 3.75LSSFCQNHH1359 pKa = 5.55FTPKK1363 pKa = 9.21STVANDD1369 pKa = 3.13YY1370 pKa = 8.79TAFDD1374 pKa = 3.61QSQLGEE1380 pKa = 4.29SLCLEE1385 pKa = 4.11VLKK1388 pKa = 10.21MKK1390 pKa = 10.5RR1391 pKa = 11.84LSIPNNFIEE1400 pKa = 5.02LHH1402 pKa = 5.47LTLKK1406 pKa = 10.76RR1407 pKa = 11.84NLYY1410 pKa = 9.88CQFGPLTCMRR1420 pKa = 11.84FTGEE1424 pKa = 3.96PGTWDD1429 pKa = 4.66DD1430 pKa = 3.68NTDD1433 pKa = 3.48YY1434 pKa = 11.21NLSVLFTLFQITNQAVLVSGDD1455 pKa = 4.04DD1456 pKa = 3.57SCINPIPPFNPRR1468 pKa = 11.84WNSIKK1473 pKa = 10.54KK1474 pKa = 10.2LLLLKK1479 pKa = 10.7FKK1481 pKa = 11.18LEE1483 pKa = 4.0YY1484 pKa = 9.89TKK1486 pKa = 10.77FPIFCGYY1493 pKa = 10.36YY1494 pKa = 9.0VGKK1497 pKa = 10.44AGALRR1502 pKa = 11.84APTALAHH1509 pKa = 6.73KK1510 pKa = 9.43IHH1512 pKa = 6.6NALADD1517 pKa = 3.86DD1518 pKa = 4.93SISDD1522 pKa = 3.34KK1523 pKa = 10.91HH1524 pKa = 7.7LSYY1527 pKa = 10.89LSEE1530 pKa = 3.92FAIGHH1535 pKa = 5.44SLGEE1539 pKa = 4.15QLWTLLPHH1547 pKa = 6.67HH1548 pKa = 6.53EE1549 pKa = 4.4VLAQSALFDD1558 pKa = 3.66YY1559 pKa = 10.3FCRR1562 pKa = 11.84KK1563 pKa = 7.68TSKK1566 pKa = 10.47KK1567 pKa = 10.36NKK1569 pKa = 8.66LLLYY1573 pKa = 9.88VGPPKK1578 pKa = 10.13QEE1580 pKa = 4.05HH1581 pKa = 6.57LNLLTFDD1588 pKa = 3.58IEE1590 pKa = 4.67SYY1592 pKa = 10.19QALRR1596 pKa = 11.84QYY1598 pKa = 10.74QRR1600 pKa = 11.84ANLLSKK1606 pKa = 10.23HH1607 pKa = 5.48QPPPLAPTLEE1617 pKa = 4.38PVLQQIFNNN1626 pKa = 3.93
MM1 pKa = 7.65SSLSNNMQILSNTLHH16 pKa = 7.02KK17 pKa = 10.71DD18 pKa = 3.19ASTHH22 pKa = 6.19PLLTHH27 pKa = 5.67TLKK30 pKa = 10.8NLHH33 pKa = 6.26TSLHH37 pKa = 5.81AFPWSLTSEE46 pKa = 4.21QVEE49 pKa = 4.15LLVNLGINVSPHH61 pKa = 4.72GTLPHH66 pKa = 6.16PHH68 pKa = 7.12PVHH71 pKa = 6.5KK72 pKa = 10.63CIEE75 pKa = 4.12THH77 pKa = 6.21VLFNHH82 pKa = 6.3WNHH85 pKa = 6.76LAVQPSAVCFMKK97 pKa = 10.38PSKK100 pKa = 9.85FAKK103 pKa = 10.19LSNSNPNFIEE113 pKa = 3.78LHH115 pKa = 6.05NYY117 pKa = 9.43IIDD120 pKa = 3.7ARR122 pKa = 11.84DD123 pKa = 3.39PTRR126 pKa = 11.84YY127 pKa = 9.35PSISSSLPTTPYY139 pKa = 11.17LFIHH143 pKa = 7.15DD144 pKa = 4.48ALMFISPNEE153 pKa = 4.19IIDD156 pKa = 4.46LFSNCPNLTHH166 pKa = 6.97VFGTLVLPAEE176 pKa = 4.94SIHH179 pKa = 6.42NIPSFYY185 pKa = 10.07PDD187 pKa = 2.93IYY189 pKa = 10.43TYY191 pKa = 10.71HH192 pKa = 6.89HH193 pKa = 6.94EE194 pKa = 4.5NGNLHH199 pKa = 6.39YY200 pKa = 9.97FLEE203 pKa = 5.26GNPSPHH209 pKa = 5.19YY210 pKa = 7.7TQPKK214 pKa = 9.34DD215 pKa = 3.14AAIWLTTHH223 pKa = 7.56TINGPNFSLSVVILEE238 pKa = 4.39SWLSVHH244 pKa = 7.05SILITKK250 pKa = 7.11YY251 pKa = 8.0QPRR254 pKa = 11.84VPTYY258 pKa = 8.99KK259 pKa = 10.25HH260 pKa = 5.71YY261 pKa = 10.78FRR263 pKa = 11.84IPKK266 pKa = 9.42TSLLPNPTNLNLPLHH281 pKa = 6.16SRR283 pKa = 11.84LVPTEE288 pKa = 3.57VHH290 pKa = 6.58ASLFNYY296 pKa = 8.49VRR298 pKa = 11.84AVRR301 pKa = 11.84TLRR304 pKa = 11.84TSDD307 pKa = 3.08PSAFIRR313 pKa = 11.84SLRR316 pKa = 11.84SKK318 pKa = 10.86EE319 pKa = 3.8EE320 pKa = 3.84YY321 pKa = 9.87RR322 pKa = 11.84WVHH325 pKa = 5.97ASAWDD330 pKa = 3.6QLVQFALATHH340 pKa = 6.44NVTSPSLFAFIEE352 pKa = 4.27TFLRR356 pKa = 11.84QILNHH361 pKa = 6.63AKK363 pKa = 9.04LTFLQFIPNPIHH375 pKa = 7.1HH376 pKa = 6.91AFFWLLAKK384 pKa = 10.16ALSYY388 pKa = 8.97ITPKK392 pKa = 10.04PYY394 pKa = 9.56STIFHH399 pKa = 5.94VASIISPLLHH409 pKa = 6.29TYY411 pKa = 8.99WLFSHH416 pKa = 7.45PSPPQVILANYY427 pKa = 7.89NQYY430 pKa = 10.45FHH432 pKa = 7.83PDD434 pKa = 3.0PWLLFADD441 pKa = 4.96RR442 pKa = 11.84YY443 pKa = 10.85SIVAMPKK450 pKa = 9.66PFLSLTLQTPPPSPTSTPPPQPTQQTQPALPAPPPVQTSNPLNSTPFAPPSSLPPLQTPQSAQPTILSTSTPVLPPIVSPPITTCNNPDD539 pKa = 4.14PLLTDD544 pKa = 3.8PTAIGVILPFNQLYY558 pKa = 9.79PGLSMPSHH566 pKa = 6.2VCNFPSRR573 pKa = 11.84LRR575 pKa = 11.84DD576 pKa = 3.44PNAPLPSYY584 pKa = 10.48PITQVCLFDD593 pKa = 4.64ALSTLLQQPQHH604 pKa = 6.19NLFQILAQYY613 pKa = 10.61LPDD616 pKa = 3.82SLVVGPEE623 pKa = 4.05EE624 pKa = 4.18KK625 pKa = 10.11TSGYY629 pKa = 8.66STDD632 pKa = 3.74HH633 pKa = 7.16LDD635 pKa = 2.94ILLYY639 pKa = 10.9HH640 pKa = 6.95FDD642 pKa = 3.55QPLIIHH648 pKa = 6.62SSLGDD653 pKa = 3.62EE654 pKa = 3.92PHH656 pKa = 6.6GPKK659 pKa = 10.16NSRR662 pKa = 11.84NLLHH666 pKa = 7.15LYY668 pKa = 10.03HH669 pKa = 6.35FTGHH673 pKa = 6.66WSAQPPSQPPLRR685 pKa = 11.84GSGLSPFANAVLSFRR700 pKa = 11.84CEE702 pKa = 4.2DD703 pKa = 3.46NSLLPIANVYY713 pKa = 6.69VHH715 pKa = 6.49KK716 pKa = 10.99VNYY719 pKa = 9.36HH720 pKa = 4.97RR721 pKa = 11.84AKK723 pKa = 10.65NLSSNLKK730 pKa = 10.1NEE732 pKa = 3.93NDD734 pKa = 4.8GILLTQLKK742 pKa = 10.34QSVNDD747 pKa = 3.51NKK749 pKa = 10.76LLSKK753 pKa = 10.2LDD755 pKa = 3.64SMMDD759 pKa = 3.54NPVSRR764 pKa = 11.84HH765 pKa = 4.74VEE767 pKa = 4.32LIHH770 pKa = 6.66IAGFPGCGKK779 pKa = 9.18SHH781 pKa = 7.09PVASLLKK788 pKa = 10.27CKK790 pKa = 7.84TFRR793 pKa = 11.84NSFRR797 pKa = 11.84VVVPTVEE804 pKa = 5.63LRR806 pKa = 11.84SEE808 pKa = 4.07WKK810 pKa = 10.42DD811 pKa = 3.36KK812 pKa = 11.72LEE814 pKa = 4.62LPDD817 pKa = 3.83FEE819 pKa = 4.53SWRR822 pKa = 11.84ISTWEE827 pKa = 3.79TSLLKK832 pKa = 10.26LAKK835 pKa = 10.26VLVIDD840 pKa = 4.86EE841 pKa = 4.58VYY843 pKa = 10.82KK844 pKa = 11.03LPNGFLDD851 pKa = 4.12LAVALDD857 pKa = 3.71PTLQFVILLGDD868 pKa = 4.14PCQGTYY874 pKa = 10.6HH875 pKa = 6.6SLSPHH880 pKa = 5.2STNHH884 pKa = 6.13HH885 pKa = 6.83IYY887 pKa = 10.41PEE889 pKa = 4.03TEE891 pKa = 4.0YY892 pKa = 10.97LSKK895 pKa = 11.18YY896 pKa = 9.93RR897 pKa = 11.84DD898 pKa = 4.6FYY900 pKa = 11.32CLWTHH905 pKa = 6.54RR906 pKa = 11.84TPQVIANLLNVRR918 pKa = 11.84SSSKK922 pKa = 10.89EE923 pKa = 3.35NGQISRR929 pKa = 11.84SNTLNASFPVLATSMAIATHH949 pKa = 6.68LNQNGFRR956 pKa = 11.84SITYY960 pKa = 9.9AGSQGLTLNRR970 pKa = 11.84LTYY973 pKa = 10.63LCVDD977 pKa = 4.27RR978 pKa = 11.84YY979 pKa = 9.9ISLMSPGVTLVALTRR994 pKa = 11.84STKK997 pKa = 10.82GIIFNGNYY1005 pKa = 10.27DD1006 pKa = 3.84LLGNLGYY1013 pKa = 9.53TNTLLEE1019 pKa = 4.54SIFRR1023 pKa = 11.84GEE1025 pKa = 4.67KK1026 pKa = 9.33IDD1028 pKa = 3.9FQHH1031 pKa = 7.23LFHH1034 pKa = 6.12QQLRR1038 pKa = 11.84GSRR1041 pKa = 11.84IITQPIMSPTDD1052 pKa = 2.99RR1053 pKa = 11.84KK1054 pKa = 9.22MLRR1057 pKa = 11.84GGAILITKK1065 pKa = 6.86TAKK1068 pKa = 9.81IDD1070 pKa = 3.49INSSIDD1076 pKa = 3.55FLRR1079 pKa = 11.84QDD1081 pKa = 3.0SQLRR1085 pKa = 11.84SNPEE1089 pKa = 4.63LIPLHH1094 pKa = 7.06DD1095 pKa = 4.56NFYY1098 pKa = 10.5LPPSRR1103 pKa = 11.84LVLSQSFSSSLHH1115 pKa = 6.27PPAKK1119 pKa = 10.17PNNTPFAPTPFEE1131 pKa = 4.02PSYY1134 pKa = 10.37PGYY1137 pKa = 10.0PFEE1140 pKa = 4.92TYY1142 pKa = 9.85FANLPDD1148 pKa = 4.23PSEE1151 pKa = 4.5PEE1153 pKa = 4.52SKK1155 pKa = 10.56EE1156 pKa = 3.33KK1157 pKa = 9.74WYY1159 pKa = 10.59KK1160 pKa = 10.57DD1161 pKa = 3.0KK1162 pKa = 10.99RR1163 pKa = 11.84SQQFPILNKK1172 pKa = 9.91GSPYY1176 pKa = 10.87SPFPANIVAPIHH1188 pKa = 6.85DD1189 pKa = 4.4SKK1191 pKa = 11.46KK1192 pKa = 10.94DD1193 pKa = 3.57STLLPLSINKK1203 pKa = 9.63RR1204 pKa = 11.84LRR1206 pKa = 11.84FRR1208 pKa = 11.84PDD1210 pKa = 2.81NKK1212 pKa = 9.99PYY1214 pKa = 10.59QITEE1218 pKa = 3.89LDD1220 pKa = 3.72HH1221 pKa = 6.72YY1222 pKa = 10.01VANHH1226 pKa = 6.85LFQSYY1231 pKa = 10.48CEE1233 pKa = 4.19TLNTTIDD1240 pKa = 3.11IKK1242 pKa = 11.45VPFDD1246 pKa = 3.07AHH1248 pKa = 7.11LFAEE1252 pKa = 5.28CINLNDD1258 pKa = 3.76YY1259 pKa = 10.56NQLTKK1264 pKa = 9.61KK1265 pKa = 8.78TRR1267 pKa = 11.84ATLVANAYY1275 pKa = 10.2RR1276 pKa = 11.84SDD1278 pKa = 3.94PDD1280 pKa = 3.0WRR1282 pKa = 11.84FTFVRR1287 pKa = 11.84IFSKK1291 pKa = 9.02TQHH1294 pKa = 6.65KK1295 pKa = 10.69INTNSLYY1302 pKa = 10.29TSWKK1306 pKa = 9.38ACQTLALMNDD1316 pKa = 4.87LIVLTLGPVKK1326 pKa = 10.46KK1327 pKa = 10.05YY1328 pKa = 10.43QRR1330 pKa = 11.84LINDD1334 pKa = 4.47PLRR1337 pKa = 11.84KK1338 pKa = 9.92SNIFTYY1344 pKa = 10.39GGKK1347 pKa = 8.06TPFDD1351 pKa = 3.75LSSFCQNHH1359 pKa = 5.55FTPKK1363 pKa = 9.21STVANDD1369 pKa = 3.13YY1370 pKa = 8.79TAFDD1374 pKa = 3.61QSQLGEE1380 pKa = 4.29SLCLEE1385 pKa = 4.11VLKK1388 pKa = 10.21MKK1390 pKa = 10.5RR1391 pKa = 11.84LSIPNNFIEE1400 pKa = 5.02LHH1402 pKa = 5.47LTLKK1406 pKa = 10.76RR1407 pKa = 11.84NLYY1410 pKa = 9.88CQFGPLTCMRR1420 pKa = 11.84FTGEE1424 pKa = 3.96PGTWDD1429 pKa = 4.66DD1430 pKa = 3.68NTDD1433 pKa = 3.48YY1434 pKa = 11.21NLSVLFTLFQITNQAVLVSGDD1455 pKa = 4.04DD1456 pKa = 3.57SCINPIPPFNPRR1468 pKa = 11.84WNSIKK1473 pKa = 10.54KK1474 pKa = 10.2LLLLKK1479 pKa = 10.7FKK1481 pKa = 11.18LEE1483 pKa = 4.0YY1484 pKa = 9.89TKK1486 pKa = 10.77FPIFCGYY1493 pKa = 10.36YY1494 pKa = 9.0VGKK1497 pKa = 10.44AGALRR1502 pKa = 11.84APTALAHH1509 pKa = 6.73KK1510 pKa = 9.43IHH1512 pKa = 6.6NALADD1517 pKa = 3.86DD1518 pKa = 4.93SISDD1522 pKa = 3.34KK1523 pKa = 10.91HH1524 pKa = 7.7LSYY1527 pKa = 10.89LSEE1530 pKa = 3.92FAIGHH1535 pKa = 5.44SLGEE1539 pKa = 4.15QLWTLLPHH1547 pKa = 6.67HH1548 pKa = 6.53EE1549 pKa = 4.4VLAQSALFDD1558 pKa = 3.66YY1559 pKa = 10.3FCRR1562 pKa = 11.84KK1563 pKa = 7.68TSKK1566 pKa = 10.47KK1567 pKa = 10.36NKK1569 pKa = 8.66LLLYY1573 pKa = 9.88VGPPKK1578 pKa = 10.13QEE1580 pKa = 4.05HH1581 pKa = 6.57LNLLTFDD1588 pKa = 3.58IEE1590 pKa = 4.67SYY1592 pKa = 10.19QALRR1596 pKa = 11.84QYY1598 pKa = 10.74QRR1600 pKa = 11.84ANLLSKK1606 pKa = 10.23HH1607 pKa = 5.48QPPPLAPTLEE1617 pKa = 4.38PVLQQIFNNN1626 pKa = 3.93
Molecular weight: 183.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1849 |
223 |
1626 |
924.5 |
104.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.408 ± 1.064 | 1.46 ± 0.312 |
4.056 ± 0.008 | 3.029 ± 0.135 |
5.138 ± 0.261 | 3.191 ± 0.158 |
4.327 ± 0.474 | 6.057 ± 1.163 |
4.651 ± 0.067 | 12.655 ± 1.473 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.973 ± 0.149 | 6.165 ± 0.672 |
9.627 ± 0.454 | 4.056 ± 0.366 |
3.407 ± 0.465 | 9.519 ± 1.213 |
6.923 ± 0.258 | 4.489 ± 0.356 |
1.19 ± 0.117 | 3.678 ± 0.573 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
