
Pohorje myodes paramyxovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Jeilongvirus; Myodes jeilongvirus
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
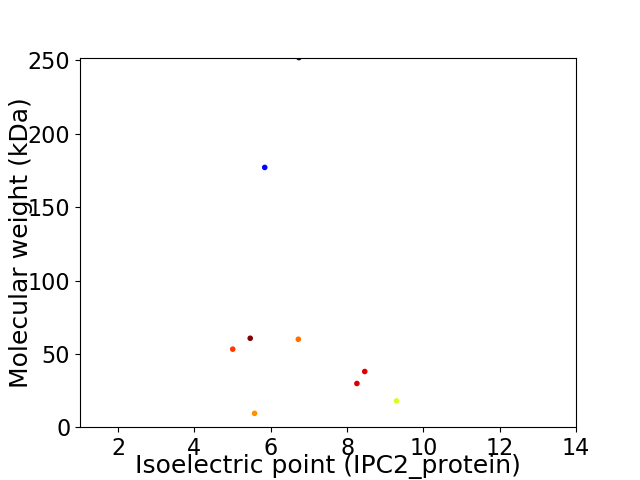
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1GJB1|A0A2P1GJB1_9MONO Nucleocapsid OS=Pohorje myodes paramyxovirus 1 OX=2116604 GN=N PE=3 SV=1
MM1 pKa = 7.62ANFSTSEE8 pKa = 4.05LNQLVQDD15 pKa = 4.82GIKK18 pKa = 9.3TVEE21 pKa = 5.21FIQQATKK28 pKa = 10.78NPKK31 pKa = 7.3EE32 pKa = 4.22TYY34 pKa = 9.78GRR36 pKa = 11.84SAIQKK41 pKa = 9.06PRR43 pKa = 11.84TKK45 pKa = 10.81DD46 pKa = 3.21RR47 pKa = 11.84IKK49 pKa = 10.38AWEE52 pKa = 4.64GVTADD57 pKa = 4.21HH58 pKa = 7.91DD59 pKa = 4.36YY60 pKa = 11.44TGDD63 pKa = 3.66KK64 pKa = 10.36QDD66 pKa = 3.8GVPGKK71 pKa = 10.48EE72 pKa = 3.64KK73 pKa = 10.6GIKK76 pKa = 8.61KK77 pKa = 10.1RR78 pKa = 11.84KK79 pKa = 9.52GKK81 pKa = 10.56GSPKK85 pKa = 10.23DD86 pKa = 3.43NRR88 pKa = 11.84GTGNEE93 pKa = 3.77EE94 pKa = 3.86DD95 pKa = 4.39PIKK98 pKa = 10.6RR99 pKa = 11.84EE100 pKa = 3.58PSTPSEE106 pKa = 4.31AGDD109 pKa = 3.39IQDD112 pKa = 3.94RR113 pKa = 11.84DD114 pKa = 3.81VEE116 pKa = 4.31NTADD120 pKa = 3.67EE121 pKa = 4.19RR122 pKa = 11.84AEE124 pKa = 4.48DD125 pKa = 4.22GGDD128 pKa = 3.34TGSGLHH134 pKa = 5.46QPRR137 pKa = 11.84EE138 pKa = 4.2GSDD141 pKa = 3.12PGSDD145 pKa = 3.4GDD147 pKa = 4.16AEE149 pKa = 4.49SARR152 pKa = 11.84SLGDD156 pKa = 3.5DD157 pKa = 3.4SVSNADD163 pKa = 3.3YY164 pKa = 10.75RR165 pKa = 11.84VILSADD171 pKa = 3.79HH172 pKa = 6.55EE173 pKa = 4.86SSAAEE178 pKa = 4.21TTDD181 pKa = 3.44LPSNVASVRR190 pKa = 11.84PATTEE195 pKa = 3.68DD196 pKa = 3.36FAQIFEE202 pKa = 4.45EE203 pKa = 5.38GSSKK207 pKa = 10.45AHH209 pKa = 6.26RR210 pKa = 11.84RR211 pKa = 11.84LTGVAAYY218 pKa = 8.1TDD220 pKa = 3.27SDD222 pKa = 4.19AAHH225 pKa = 7.04KK226 pKa = 10.79SAGNPVKK233 pKa = 10.6KK234 pKa = 10.7GIDD237 pKa = 3.3GSTASTPSVDD247 pKa = 3.31IQSSGSGAIPSVHH260 pKa = 6.42EE261 pKa = 4.06LLLRR265 pKa = 11.84QPSSHH270 pKa = 5.55AHH272 pKa = 6.16AEE274 pKa = 4.34SAHH277 pKa = 6.3KK278 pKa = 10.3GVPSVSTTGSTYY290 pKa = 10.63KK291 pKa = 10.36SCEE294 pKa = 4.18TASHH298 pKa = 5.59HH299 pKa = 6.67QILEE303 pKa = 4.0GKK305 pKa = 9.8IDD307 pKa = 3.62ILIGNVDD314 pKa = 4.98RR315 pKa = 11.84IASKK319 pKa = 11.11LDD321 pKa = 3.48LLPEE325 pKa = 3.9IKK327 pKa = 10.6EE328 pKa = 4.04EE329 pKa = 3.86IKK331 pKa = 10.99NINKK335 pKa = 10.31KK336 pKa = 8.24ITTLSLGLSTVEE348 pKa = 4.21NYY350 pKa = 9.38IKK352 pKa = 11.04SMMVIIPGSGKK363 pKa = 10.2DD364 pKa = 3.47EE365 pKa = 4.4EE366 pKa = 4.71KK367 pKa = 10.75RR368 pKa = 11.84NPDD371 pKa = 3.24VNPDD375 pKa = 3.45LRR377 pKa = 11.84PVIGRR382 pKa = 11.84DD383 pKa = 3.0NTRR386 pKa = 11.84GLRR389 pKa = 11.84EE390 pKa = 3.88MKK392 pKa = 9.64MEE394 pKa = 4.98RR395 pKa = 11.84GTLEE399 pKa = 4.37DD400 pKa = 5.75LNDD403 pKa = 3.83EE404 pKa = 4.89SNSQPKK410 pKa = 9.49IDD412 pKa = 4.35PKK414 pKa = 11.35YY415 pKa = 10.6LINPLDD421 pKa = 3.89FSKK424 pKa = 11.45SNAANFKK431 pKa = 8.26PTNDD435 pKa = 3.55LASLKK440 pKa = 10.01TIVAMIKK447 pKa = 10.53NEE449 pKa = 4.05VKK451 pKa = 10.67DD452 pKa = 3.17ITTQISLIEE461 pKa = 4.07WVEE464 pKa = 3.93KK465 pKa = 10.83KK466 pKa = 9.88IDD468 pKa = 3.56QLPAEE473 pKa = 4.48EE474 pKa = 4.87VYY476 pKa = 11.62NMVRR480 pKa = 11.84EE481 pKa = 4.34SLDD484 pKa = 3.42GMDD487 pKa = 4.2SDD489 pKa = 4.83EE490 pKa = 4.28SS491 pKa = 3.76
MM1 pKa = 7.62ANFSTSEE8 pKa = 4.05LNQLVQDD15 pKa = 4.82GIKK18 pKa = 9.3TVEE21 pKa = 5.21FIQQATKK28 pKa = 10.78NPKK31 pKa = 7.3EE32 pKa = 4.22TYY34 pKa = 9.78GRR36 pKa = 11.84SAIQKK41 pKa = 9.06PRR43 pKa = 11.84TKK45 pKa = 10.81DD46 pKa = 3.21RR47 pKa = 11.84IKK49 pKa = 10.38AWEE52 pKa = 4.64GVTADD57 pKa = 4.21HH58 pKa = 7.91DD59 pKa = 4.36YY60 pKa = 11.44TGDD63 pKa = 3.66KK64 pKa = 10.36QDD66 pKa = 3.8GVPGKK71 pKa = 10.48EE72 pKa = 3.64KK73 pKa = 10.6GIKK76 pKa = 8.61KK77 pKa = 10.1RR78 pKa = 11.84KK79 pKa = 9.52GKK81 pKa = 10.56GSPKK85 pKa = 10.23DD86 pKa = 3.43NRR88 pKa = 11.84GTGNEE93 pKa = 3.77EE94 pKa = 3.86DD95 pKa = 4.39PIKK98 pKa = 10.6RR99 pKa = 11.84EE100 pKa = 3.58PSTPSEE106 pKa = 4.31AGDD109 pKa = 3.39IQDD112 pKa = 3.94RR113 pKa = 11.84DD114 pKa = 3.81VEE116 pKa = 4.31NTADD120 pKa = 3.67EE121 pKa = 4.19RR122 pKa = 11.84AEE124 pKa = 4.48DD125 pKa = 4.22GGDD128 pKa = 3.34TGSGLHH134 pKa = 5.46QPRR137 pKa = 11.84EE138 pKa = 4.2GSDD141 pKa = 3.12PGSDD145 pKa = 3.4GDD147 pKa = 4.16AEE149 pKa = 4.49SARR152 pKa = 11.84SLGDD156 pKa = 3.5DD157 pKa = 3.4SVSNADD163 pKa = 3.3YY164 pKa = 10.75RR165 pKa = 11.84VILSADD171 pKa = 3.79HH172 pKa = 6.55EE173 pKa = 4.86SSAAEE178 pKa = 4.21TTDD181 pKa = 3.44LPSNVASVRR190 pKa = 11.84PATTEE195 pKa = 3.68DD196 pKa = 3.36FAQIFEE202 pKa = 4.45EE203 pKa = 5.38GSSKK207 pKa = 10.45AHH209 pKa = 6.26RR210 pKa = 11.84RR211 pKa = 11.84LTGVAAYY218 pKa = 8.1TDD220 pKa = 3.27SDD222 pKa = 4.19AAHH225 pKa = 7.04KK226 pKa = 10.79SAGNPVKK233 pKa = 10.6KK234 pKa = 10.7GIDD237 pKa = 3.3GSTASTPSVDD247 pKa = 3.31IQSSGSGAIPSVHH260 pKa = 6.42EE261 pKa = 4.06LLLRR265 pKa = 11.84QPSSHH270 pKa = 5.55AHH272 pKa = 6.16AEE274 pKa = 4.34SAHH277 pKa = 6.3KK278 pKa = 10.3GVPSVSTTGSTYY290 pKa = 10.63KK291 pKa = 10.36SCEE294 pKa = 4.18TASHH298 pKa = 5.59HH299 pKa = 6.67QILEE303 pKa = 4.0GKK305 pKa = 9.8IDD307 pKa = 3.62ILIGNVDD314 pKa = 4.98RR315 pKa = 11.84IASKK319 pKa = 11.11LDD321 pKa = 3.48LLPEE325 pKa = 3.9IKK327 pKa = 10.6EE328 pKa = 4.04EE329 pKa = 3.86IKK331 pKa = 10.99NINKK335 pKa = 10.31KK336 pKa = 8.24ITTLSLGLSTVEE348 pKa = 4.21NYY350 pKa = 9.38IKK352 pKa = 11.04SMMVIIPGSGKK363 pKa = 10.2DD364 pKa = 3.47EE365 pKa = 4.4EE366 pKa = 4.71KK367 pKa = 10.75RR368 pKa = 11.84NPDD371 pKa = 3.24VNPDD375 pKa = 3.45LRR377 pKa = 11.84PVIGRR382 pKa = 11.84DD383 pKa = 3.0NTRR386 pKa = 11.84GLRR389 pKa = 11.84EE390 pKa = 3.88MKK392 pKa = 9.64MEE394 pKa = 4.98RR395 pKa = 11.84GTLEE399 pKa = 4.37DD400 pKa = 5.75LNDD403 pKa = 3.83EE404 pKa = 4.89SNSQPKK410 pKa = 9.49IDD412 pKa = 4.35PKK414 pKa = 11.35YY415 pKa = 10.6LINPLDD421 pKa = 3.89FSKK424 pKa = 11.45SNAANFKK431 pKa = 8.26PTNDD435 pKa = 3.55LASLKK440 pKa = 10.01TIVAMIKK447 pKa = 10.53NEE449 pKa = 4.05VKK451 pKa = 10.67DD452 pKa = 3.17ITTQISLIEE461 pKa = 4.07WVEE464 pKa = 3.93KK465 pKa = 10.83KK466 pKa = 9.88IDD468 pKa = 3.56QLPAEE473 pKa = 4.48EE474 pKa = 4.87VYY476 pKa = 11.62NMVRR480 pKa = 11.84EE481 pKa = 4.34SLDD484 pKa = 3.42GMDD487 pKa = 4.2SDD489 pKa = 4.83EE490 pKa = 4.28SS491 pKa = 3.76
Molecular weight: 53.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1GJA8|A0A2P1GJA8_9MONO TM protein OS=Pohorje myodes paramyxovirus 1 OX=2116604 GN=TM PE=4 SV=1
MM1 pKa = 7.42EE2 pKa = 5.62SKK4 pKa = 10.73LSSLYY9 pKa = 10.19NRR11 pKa = 11.84LRR13 pKa = 11.84RR14 pKa = 11.84TLRR17 pKa = 11.84KK18 pKa = 8.45HH19 pKa = 5.02TGEE22 pKa = 4.06VPSRR26 pKa = 11.84NQEE29 pKa = 3.67PRR31 pKa = 11.84TEE33 pKa = 4.02SRR35 pKa = 11.84PGRR38 pKa = 11.84EE39 pKa = 4.21SQQTMITQEE48 pKa = 4.22TNRR51 pKa = 11.84MEE53 pKa = 4.36CLEE56 pKa = 4.01RR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84EE60 pKa = 4.03LRR62 pKa = 11.84KK63 pKa = 9.84EE64 pKa = 3.51RR65 pKa = 11.84AKK67 pKa = 10.55EE68 pKa = 3.53VLRR71 pKa = 11.84IIEE74 pKa = 4.3EE75 pKa = 4.42LEE77 pKa = 3.89MKK79 pKa = 10.51RR80 pKa = 11.84ILSRR84 pKa = 11.84EE85 pKa = 4.02NPQPPPRR92 pKa = 11.84LAISRR97 pKa = 11.84TGMLKK102 pKa = 9.82ILLMSVQKK110 pKa = 9.82TGEE113 pKa = 4.22IPVLDD118 pKa = 4.42YY119 pKa = 11.17ISLEE123 pKa = 3.88RR124 pKa = 11.84AQILDD129 pKa = 3.49QMEE132 pKa = 4.21MQSLRR137 pKa = 11.84EE138 pKa = 4.2AWVMIQSVMRR148 pKa = 11.84ITVV151 pKa = 2.94
MM1 pKa = 7.42EE2 pKa = 5.62SKK4 pKa = 10.73LSSLYY9 pKa = 10.19NRR11 pKa = 11.84LRR13 pKa = 11.84RR14 pKa = 11.84TLRR17 pKa = 11.84KK18 pKa = 8.45HH19 pKa = 5.02TGEE22 pKa = 4.06VPSRR26 pKa = 11.84NQEE29 pKa = 3.67PRR31 pKa = 11.84TEE33 pKa = 4.02SRR35 pKa = 11.84PGRR38 pKa = 11.84EE39 pKa = 4.21SQQTMITQEE48 pKa = 4.22TNRR51 pKa = 11.84MEE53 pKa = 4.36CLEE56 pKa = 4.01RR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84EE60 pKa = 4.03LRR62 pKa = 11.84KK63 pKa = 9.84EE64 pKa = 3.51RR65 pKa = 11.84AKK67 pKa = 10.55EE68 pKa = 3.53VLRR71 pKa = 11.84IIEE74 pKa = 4.3EE75 pKa = 4.42LEE77 pKa = 3.89MKK79 pKa = 10.51RR80 pKa = 11.84ILSRR84 pKa = 11.84EE85 pKa = 4.02NPQPPPRR92 pKa = 11.84LAISRR97 pKa = 11.84TGMLKK102 pKa = 9.82ILLMSVQKK110 pKa = 9.82TGEE113 pKa = 4.22IPVLDD118 pKa = 4.42YY119 pKa = 11.17ISLEE123 pKa = 3.88RR124 pKa = 11.84AQILDD129 pKa = 3.49QMEE132 pKa = 4.21MQSLRR137 pKa = 11.84EE138 pKa = 4.2AWVMIQSVMRR148 pKa = 11.84ITVV151 pKa = 2.94
Molecular weight: 17.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6189 |
82 |
2188 |
687.7 |
77.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.381 ± 0.651 | 1.697 ± 0.325 |
5.865 ± 0.545 | 5.494 ± 0.388 |
3.442 ± 0.4 | 5.526 ± 0.454 |
2.068 ± 0.345 | 7.513 ± 0.576 |
5.704 ± 0.529 | 8.838 ± 0.874 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.908 ± 0.356 | 5.93 ± 0.166 |
5.251 ± 0.957 | 3.781 ± 0.256 |
5.251 ± 0.349 | 8.014 ± 0.52 |
7.465 ± 1.225 | 4.863 ± 0.465 |
0.969 ± 0.151 | 4.039 ± 0.524 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
