
Antonospora locustae virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Amalgaviridae; unclassified Amalgaviridae
Average proteome isoelectric point is 7.8
Get precalculated fractions of proteins
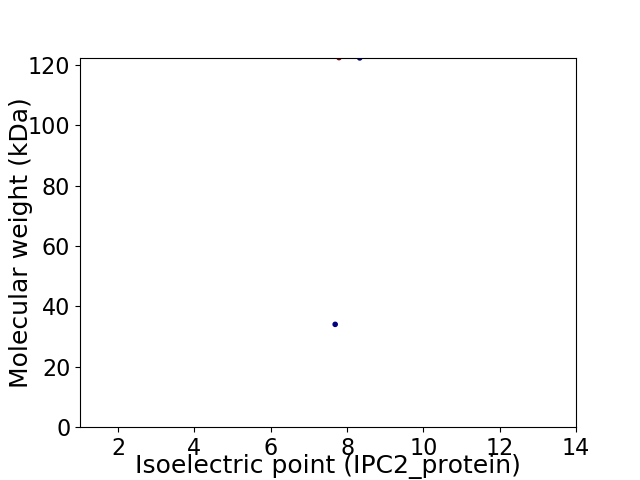
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A218KIR8|A0A218KIR8_9VIRU ORF1/ORF2 fusion protein OS=Antonospora locustae virus 1 OX=1872708 PE=4 SV=1
MM1 pKa = 7.02EE2 pKa = 5.45ASVFVSRR9 pKa = 11.84LMAIEE14 pKa = 4.58PSDD17 pKa = 3.91FSQHH21 pKa = 5.43FSLPMSICDD30 pKa = 4.17HH31 pKa = 5.8GLKK34 pKa = 9.52MIRR37 pKa = 11.84ISAEE41 pKa = 3.46QTPAFVKK48 pKa = 10.82AFTTQYY54 pKa = 11.14ALSNEE59 pKa = 4.33CPQTEE64 pKa = 4.39FFNALFDD71 pKa = 4.46YY72 pKa = 11.23IMDD75 pKa = 4.24GKK77 pKa = 10.14IKK79 pKa = 10.19QAIGEE84 pKa = 4.03ASGRR88 pKa = 11.84LRR90 pKa = 11.84KK91 pKa = 8.29EE92 pKa = 3.63QKK94 pKa = 10.26LLQFQEE100 pKa = 3.89LHH102 pKa = 6.28GFSKK106 pKa = 10.75DD107 pKa = 3.23IEE109 pKa = 4.67DD110 pKa = 4.91DD111 pKa = 3.87FNEE114 pKa = 4.99ALILYY119 pKa = 8.83NKK121 pKa = 9.02EE122 pKa = 3.83RR123 pKa = 11.84ADD125 pKa = 3.69LRR127 pKa = 11.84SVKK130 pKa = 10.66DD131 pKa = 3.66KK132 pKa = 10.97FDD134 pKa = 3.98EE135 pKa = 4.26LTKK138 pKa = 10.79DD139 pKa = 3.16LNGIFKK145 pKa = 10.63VLEE148 pKa = 4.19EE149 pKa = 4.4EE150 pKa = 5.16LSQRR154 pKa = 11.84WSALEE159 pKa = 3.85TIINEE164 pKa = 4.36KK165 pKa = 8.33KK166 pKa = 8.96TNAAEE171 pKa = 3.95AQKK174 pKa = 10.36RR175 pKa = 11.84ARR177 pKa = 11.84TNWINMKK184 pKa = 10.12QADD187 pKa = 4.33RR188 pKa = 11.84QKK190 pKa = 10.82YY191 pKa = 9.71SNDD194 pKa = 2.35FMLYY198 pKa = 9.71LKK200 pKa = 10.3EE201 pKa = 3.68YY202 pKa = 10.16SLRR205 pKa = 11.84NKK207 pKa = 10.11VNFDD211 pKa = 3.32QKK213 pKa = 11.11LKK215 pKa = 10.8SNEE218 pKa = 3.89DD219 pKa = 3.23FKK221 pKa = 11.77KK222 pKa = 10.33KK223 pKa = 10.48LKK225 pKa = 10.18EE226 pKa = 3.71LLVKK230 pKa = 10.31IRR232 pKa = 11.84VEE234 pKa = 4.08EE235 pKa = 4.01NVDD238 pKa = 4.04KK239 pKa = 11.15IFLADD244 pKa = 3.59GLINPKK250 pKa = 9.96FKK252 pKa = 10.78SEE254 pKa = 3.72RR255 pKa = 11.84VTFLVSEE262 pKa = 4.37LFEE265 pKa = 4.32KK266 pKa = 10.76LGSIKK271 pKa = 10.49KK272 pKa = 9.76IFTSQRR278 pKa = 11.84INRR281 pKa = 11.84QSGVQEE287 pKa = 4.06CTAGNLL293 pKa = 3.54
MM1 pKa = 7.02EE2 pKa = 5.45ASVFVSRR9 pKa = 11.84LMAIEE14 pKa = 4.58PSDD17 pKa = 3.91FSQHH21 pKa = 5.43FSLPMSICDD30 pKa = 4.17HH31 pKa = 5.8GLKK34 pKa = 9.52MIRR37 pKa = 11.84ISAEE41 pKa = 3.46QTPAFVKK48 pKa = 10.82AFTTQYY54 pKa = 11.14ALSNEE59 pKa = 4.33CPQTEE64 pKa = 4.39FFNALFDD71 pKa = 4.46YY72 pKa = 11.23IMDD75 pKa = 4.24GKK77 pKa = 10.14IKK79 pKa = 10.19QAIGEE84 pKa = 4.03ASGRR88 pKa = 11.84LRR90 pKa = 11.84KK91 pKa = 8.29EE92 pKa = 3.63QKK94 pKa = 10.26LLQFQEE100 pKa = 3.89LHH102 pKa = 6.28GFSKK106 pKa = 10.75DD107 pKa = 3.23IEE109 pKa = 4.67DD110 pKa = 4.91DD111 pKa = 3.87FNEE114 pKa = 4.99ALILYY119 pKa = 8.83NKK121 pKa = 9.02EE122 pKa = 3.83RR123 pKa = 11.84ADD125 pKa = 3.69LRR127 pKa = 11.84SVKK130 pKa = 10.66DD131 pKa = 3.66KK132 pKa = 10.97FDD134 pKa = 3.98EE135 pKa = 4.26LTKK138 pKa = 10.79DD139 pKa = 3.16LNGIFKK145 pKa = 10.63VLEE148 pKa = 4.19EE149 pKa = 4.4EE150 pKa = 5.16LSQRR154 pKa = 11.84WSALEE159 pKa = 3.85TIINEE164 pKa = 4.36KK165 pKa = 8.33KK166 pKa = 8.96TNAAEE171 pKa = 3.95AQKK174 pKa = 10.36RR175 pKa = 11.84ARR177 pKa = 11.84TNWINMKK184 pKa = 10.12QADD187 pKa = 4.33RR188 pKa = 11.84QKK190 pKa = 10.82YY191 pKa = 9.71SNDD194 pKa = 2.35FMLYY198 pKa = 9.71LKK200 pKa = 10.3EE201 pKa = 3.68YY202 pKa = 10.16SLRR205 pKa = 11.84NKK207 pKa = 10.11VNFDD211 pKa = 3.32QKK213 pKa = 11.11LKK215 pKa = 10.8SNEE218 pKa = 3.89DD219 pKa = 3.23FKK221 pKa = 11.77KK222 pKa = 10.33KK223 pKa = 10.48LKK225 pKa = 10.18EE226 pKa = 3.71LLVKK230 pKa = 10.31IRR232 pKa = 11.84VEE234 pKa = 4.08EE235 pKa = 4.01NVDD238 pKa = 4.04KK239 pKa = 11.15IFLADD244 pKa = 3.59GLINPKK250 pKa = 9.96FKK252 pKa = 10.78SEE254 pKa = 3.72RR255 pKa = 11.84VTFLVSEE262 pKa = 4.37LFEE265 pKa = 4.32KK266 pKa = 10.76LGSIKK271 pKa = 10.49KK272 pKa = 9.76IFTSQRR278 pKa = 11.84INRR281 pKa = 11.84QSGVQEE287 pKa = 4.06CTAGNLL293 pKa = 3.54
Molecular weight: 34.06 kDa
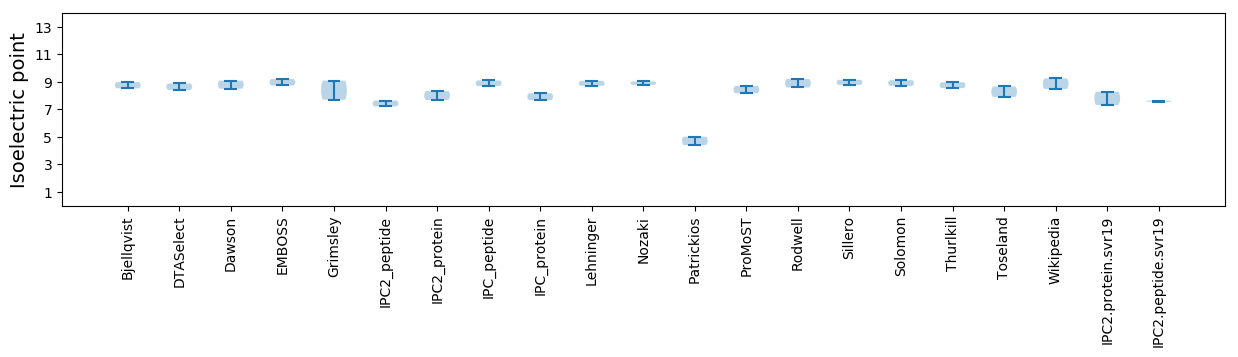
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A218KIR8|A0A218KIR8_9VIRU ORF1/ORF2 fusion protein OS=Antonospora locustae virus 1 OX=1872708 PE=4 SV=1
MM1 pKa = 7.02EE2 pKa = 5.45ASVFVSRR9 pKa = 11.84LMAIEE14 pKa = 4.58PSDD17 pKa = 3.91FSQHH21 pKa = 5.43FSLPMSICDD30 pKa = 4.17HH31 pKa = 5.8GLKK34 pKa = 9.52MIRR37 pKa = 11.84ISAEE41 pKa = 3.46QTPAFVKK48 pKa = 10.82AFTTQYY54 pKa = 11.14ALSNEE59 pKa = 4.33CPQTEE64 pKa = 4.39FFNALFDD71 pKa = 4.46YY72 pKa = 11.23IMDD75 pKa = 4.24GKK77 pKa = 10.14IKK79 pKa = 10.19QAIGEE84 pKa = 4.03ASGRR88 pKa = 11.84LRR90 pKa = 11.84KK91 pKa = 8.29EE92 pKa = 3.63QKK94 pKa = 10.26LLQFQEE100 pKa = 3.89LHH102 pKa = 6.28GFSKK106 pKa = 10.75DD107 pKa = 3.23IEE109 pKa = 4.67DD110 pKa = 4.91DD111 pKa = 3.87FNEE114 pKa = 4.99ALILYY119 pKa = 8.83NKK121 pKa = 9.02EE122 pKa = 3.83RR123 pKa = 11.84ADD125 pKa = 3.69LRR127 pKa = 11.84SVKK130 pKa = 10.66DD131 pKa = 3.66KK132 pKa = 10.97FDD134 pKa = 3.98EE135 pKa = 4.26LTKK138 pKa = 10.79DD139 pKa = 3.16LNGIFKK145 pKa = 10.63VLEE148 pKa = 4.19EE149 pKa = 4.4EE150 pKa = 5.16LSQRR154 pKa = 11.84WSALEE159 pKa = 3.85TIINEE164 pKa = 4.36KK165 pKa = 8.33KK166 pKa = 8.96TNAAEE171 pKa = 3.95AQKK174 pKa = 10.36RR175 pKa = 11.84ARR177 pKa = 11.84TNWINMKK184 pKa = 10.12QADD187 pKa = 4.33RR188 pKa = 11.84QKK190 pKa = 10.82YY191 pKa = 9.71SNDD194 pKa = 2.35FMLYY198 pKa = 9.71LKK200 pKa = 10.3EE201 pKa = 3.68YY202 pKa = 10.16SLRR205 pKa = 11.84NKK207 pKa = 10.11VNFDD211 pKa = 3.32QKK213 pKa = 11.11LKK215 pKa = 10.8SNEE218 pKa = 3.89DD219 pKa = 3.23FKK221 pKa = 11.77KK222 pKa = 10.33KK223 pKa = 10.48LKK225 pKa = 10.18EE226 pKa = 3.71LLVKK230 pKa = 10.31IRR232 pKa = 11.84VEE234 pKa = 4.08EE235 pKa = 4.01NVDD238 pKa = 4.04KK239 pKa = 11.15IFLADD244 pKa = 3.59GLINPKK250 pKa = 9.96FKK252 pKa = 10.78SEE254 pKa = 3.77RR255 pKa = 11.84VTFLSQSYY263 pKa = 10.07LRR265 pKa = 11.84NLDD268 pKa = 3.26RR269 pKa = 11.84LRR271 pKa = 11.84RR272 pKa = 11.84SLQARR277 pKa = 11.84GLTDD281 pKa = 3.19SQEE284 pKa = 4.29YY285 pKa = 10.95KK286 pKa = 10.84NVLQAIYY293 pKa = 10.83DD294 pKa = 3.72LLYY297 pKa = 11.0NFNGPYY303 pKa = 9.96LHH305 pKa = 7.65PDD307 pKa = 3.55FPGDD311 pKa = 3.41FGHH314 pKa = 7.58RR315 pKa = 11.84SHH317 pKa = 7.07ALANSFINPPLFSVPEE333 pKa = 3.87QLDD336 pKa = 3.82ANVFTSDD343 pKa = 3.35CVSNGRR349 pKa = 11.84YY350 pKa = 8.95FKK352 pKa = 11.13YY353 pKa = 10.89NEE355 pKa = 4.03FLRR358 pKa = 11.84DD359 pKa = 3.41SLGAIGGGANDD370 pKa = 4.44LLRR373 pKa = 11.84EE374 pKa = 3.95LMEE377 pKa = 3.73YY378 pKa = 10.31RR379 pKa = 11.84RR380 pKa = 11.84LGGSTSDD387 pKa = 3.18KK388 pKa = 10.97LNYY391 pKa = 9.49LAYY394 pKa = 10.37FGNKK398 pKa = 7.6SQCDD402 pKa = 3.6TLLGLDD408 pKa = 3.69TAPVYY413 pKa = 11.5SMFVSGVVRR422 pKa = 11.84EE423 pKa = 4.8PYY425 pKa = 10.12VHH427 pKa = 6.23MPYY430 pKa = 10.55SVLVKK435 pKa = 10.48KK436 pKa = 8.42FTYY439 pKa = 9.89NAQASGGTLDD449 pKa = 3.88KK450 pKa = 11.41QFGFKK455 pKa = 10.42NKK457 pKa = 10.22SDD459 pKa = 3.78VLLLNLVAYY468 pKa = 10.16RR469 pKa = 11.84LISTAWCIQKK479 pKa = 10.2RR480 pKa = 11.84SKK482 pKa = 10.58LKK484 pKa = 10.49PGIIWDD490 pKa = 3.68SASRR494 pKa = 11.84PKK496 pKa = 10.4LIKK499 pKa = 10.14IAGTGNKK506 pKa = 8.05MKK508 pKa = 10.36KK509 pKa = 9.78IMAGEE514 pKa = 4.11PCCRR518 pKa = 11.84VIAVGPLLEE527 pKa = 4.42SLLGYY532 pKa = 9.97SLYY535 pKa = 10.91LRR537 pKa = 11.84VSEE540 pKa = 5.13RR541 pKa = 11.84LQQFFKK547 pKa = 10.89EE548 pKa = 4.0KK549 pKa = 10.31GYY551 pKa = 11.13GVAIGCNRR559 pKa = 11.84MGKK562 pKa = 9.4DD563 pKa = 2.85WQNIQKK569 pKa = 9.33NFKK572 pKa = 8.69GARR575 pKa = 11.84HH576 pKa = 5.72IMVGDD581 pKa = 3.45YY582 pKa = 10.92SKK584 pKa = 11.52YY585 pKa = 10.39DD586 pKa = 3.18QTIPEE591 pKa = 4.12RR592 pKa = 11.84LMIFGIDD599 pKa = 3.97MILNMYY605 pKa = 8.49MPQDD609 pKa = 4.34EE610 pKa = 4.59YY611 pKa = 11.84SMNYY615 pKa = 9.02IDD617 pKa = 6.17NFRR620 pKa = 11.84TWFIDD625 pKa = 3.59NIVRR629 pKa = 11.84SVHH632 pKa = 5.71VVDD635 pKa = 5.28GKK637 pKa = 11.11LSARR641 pKa = 11.84AIGGMPSGTLWTSLLNSVINIVVISDD667 pKa = 3.4TCKK670 pKa = 10.45ALGIKK675 pKa = 9.77NYY677 pKa = 10.28KK678 pKa = 8.44PVVYY682 pKa = 10.11GDD684 pKa = 3.45DD685 pKa = 3.56HH686 pKa = 6.69MIIIYY691 pKa = 10.1DD692 pKa = 3.99EE693 pKa = 4.44IVDD696 pKa = 3.68SGEE699 pKa = 4.25FISAYY704 pKa = 7.91STYY707 pKa = 11.03VKK709 pKa = 10.38INFGMTLTTDD719 pKa = 3.35DD720 pKa = 4.46TYY722 pKa = 11.38ISGPEE727 pKa = 4.09CFYY730 pKa = 10.52VTYY733 pKa = 10.27KK734 pKa = 10.71RR735 pKa = 11.84PVYY738 pKa = 10.23NPKK741 pKa = 10.51ADD743 pKa = 3.71LQKK746 pKa = 9.53GTRR749 pKa = 11.84RR750 pKa = 11.84LRR752 pKa = 11.84PKK754 pKa = 10.08SWEE757 pKa = 3.87TSKK760 pKa = 9.95TPFTQFDD767 pKa = 3.68HH768 pKa = 6.71TKK770 pKa = 9.03GTTHH774 pKa = 6.47RR775 pKa = 11.84WSYY778 pKa = 9.02EE779 pKa = 3.25FRR781 pKa = 11.84GRR783 pKa = 11.84PKK785 pKa = 10.21FLQYY789 pKa = 11.01YY790 pKa = 8.1WLEE793 pKa = 3.97TGLAIRR799 pKa = 11.84PLRR802 pKa = 11.84EE803 pKa = 3.69SMVRR807 pKa = 11.84MLHH810 pKa = 6.52PEE812 pKa = 3.93EE813 pKa = 4.01EE814 pKa = 4.51VKK816 pKa = 10.66NVRR819 pKa = 11.84EE820 pKa = 3.98YY821 pKa = 9.65EE822 pKa = 4.27VLVISHH828 pKa = 7.32LYY830 pKa = 10.34DD831 pKa = 3.86NYY833 pKa = 11.1HH834 pKa = 5.99NAHH837 pKa = 5.2MRR839 pKa = 11.84NWAFHH844 pKa = 6.59LLYY847 pKa = 10.7DD848 pKa = 3.53IDD850 pKa = 3.89FMKK853 pKa = 10.42RR854 pKa = 11.84ARR856 pKa = 11.84MKK858 pKa = 10.79SADD861 pKa = 3.84LFFKK865 pKa = 10.8RR866 pKa = 11.84SDD868 pKa = 3.37RR869 pKa = 11.84GKK871 pKa = 10.92GYY873 pKa = 10.51YY874 pKa = 8.53EE875 pKa = 4.3HH876 pKa = 7.17NKK878 pKa = 9.84GPPSRR883 pKa = 11.84MWYY886 pKa = 9.84RR887 pKa = 11.84RR888 pKa = 11.84VDD890 pKa = 3.59YY891 pKa = 10.93LVDD894 pKa = 4.19LDD896 pKa = 4.25SCVGMQHH903 pKa = 6.54FVHH906 pKa = 5.85SWKK909 pKa = 10.75VLLAQMDD916 pKa = 4.26EE917 pKa = 4.75IISSEE922 pKa = 4.34HH923 pKa = 5.35EE924 pKa = 4.03CEE926 pKa = 4.37PYY928 pKa = 10.0QVRR931 pKa = 11.84HH932 pKa = 6.36LIKK935 pKa = 10.65NEE937 pKa = 3.24LSMRR941 pKa = 11.84RR942 pKa = 11.84ISTTRR947 pKa = 11.84ARR949 pKa = 11.84AWFEE953 pKa = 3.43AGIITRR959 pKa = 11.84SEE961 pKa = 4.06YY962 pKa = 11.15AQFLAAEE969 pKa = 4.64GTSKK973 pKa = 10.42QFEE976 pKa = 4.72SKK978 pKa = 10.43KK979 pKa = 10.8SPFNLFGLDD988 pKa = 3.29EE989 pKa = 4.37VGFNYY994 pKa = 10.31LFSLLGISNDD1004 pKa = 3.12KK1005 pKa = 10.18VHH1007 pKa = 6.68YY1008 pKa = 9.25STDD1011 pKa = 3.47YY1012 pKa = 11.35DD1013 pKa = 3.67RR1014 pKa = 11.84GPSRR1018 pKa = 11.84LKK1020 pKa = 10.42QVLATSIDD1028 pKa = 3.98LNGFASVVNFASKK1041 pKa = 9.77WLQSSDD1047 pKa = 3.43SPNLYY1052 pKa = 8.81RR1053 pKa = 11.84TGEE1056 pKa = 4.08PPP1058 pKa = 3.37
MM1 pKa = 7.02EE2 pKa = 5.45ASVFVSRR9 pKa = 11.84LMAIEE14 pKa = 4.58PSDD17 pKa = 3.91FSQHH21 pKa = 5.43FSLPMSICDD30 pKa = 4.17HH31 pKa = 5.8GLKK34 pKa = 9.52MIRR37 pKa = 11.84ISAEE41 pKa = 3.46QTPAFVKK48 pKa = 10.82AFTTQYY54 pKa = 11.14ALSNEE59 pKa = 4.33CPQTEE64 pKa = 4.39FFNALFDD71 pKa = 4.46YY72 pKa = 11.23IMDD75 pKa = 4.24GKK77 pKa = 10.14IKK79 pKa = 10.19QAIGEE84 pKa = 4.03ASGRR88 pKa = 11.84LRR90 pKa = 11.84KK91 pKa = 8.29EE92 pKa = 3.63QKK94 pKa = 10.26LLQFQEE100 pKa = 3.89LHH102 pKa = 6.28GFSKK106 pKa = 10.75DD107 pKa = 3.23IEE109 pKa = 4.67DD110 pKa = 4.91DD111 pKa = 3.87FNEE114 pKa = 4.99ALILYY119 pKa = 8.83NKK121 pKa = 9.02EE122 pKa = 3.83RR123 pKa = 11.84ADD125 pKa = 3.69LRR127 pKa = 11.84SVKK130 pKa = 10.66DD131 pKa = 3.66KK132 pKa = 10.97FDD134 pKa = 3.98EE135 pKa = 4.26LTKK138 pKa = 10.79DD139 pKa = 3.16LNGIFKK145 pKa = 10.63VLEE148 pKa = 4.19EE149 pKa = 4.4EE150 pKa = 5.16LSQRR154 pKa = 11.84WSALEE159 pKa = 3.85TIINEE164 pKa = 4.36KK165 pKa = 8.33KK166 pKa = 8.96TNAAEE171 pKa = 3.95AQKK174 pKa = 10.36RR175 pKa = 11.84ARR177 pKa = 11.84TNWINMKK184 pKa = 10.12QADD187 pKa = 4.33RR188 pKa = 11.84QKK190 pKa = 10.82YY191 pKa = 9.71SNDD194 pKa = 2.35FMLYY198 pKa = 9.71LKK200 pKa = 10.3EE201 pKa = 3.68YY202 pKa = 10.16SLRR205 pKa = 11.84NKK207 pKa = 10.11VNFDD211 pKa = 3.32QKK213 pKa = 11.11LKK215 pKa = 10.8SNEE218 pKa = 3.89DD219 pKa = 3.23FKK221 pKa = 11.77KK222 pKa = 10.33KK223 pKa = 10.48LKK225 pKa = 10.18EE226 pKa = 3.71LLVKK230 pKa = 10.31IRR232 pKa = 11.84VEE234 pKa = 4.08EE235 pKa = 4.01NVDD238 pKa = 4.04KK239 pKa = 11.15IFLADD244 pKa = 3.59GLINPKK250 pKa = 9.96FKK252 pKa = 10.78SEE254 pKa = 3.77RR255 pKa = 11.84VTFLSQSYY263 pKa = 10.07LRR265 pKa = 11.84NLDD268 pKa = 3.26RR269 pKa = 11.84LRR271 pKa = 11.84RR272 pKa = 11.84SLQARR277 pKa = 11.84GLTDD281 pKa = 3.19SQEE284 pKa = 4.29YY285 pKa = 10.95KK286 pKa = 10.84NVLQAIYY293 pKa = 10.83DD294 pKa = 3.72LLYY297 pKa = 11.0NFNGPYY303 pKa = 9.96LHH305 pKa = 7.65PDD307 pKa = 3.55FPGDD311 pKa = 3.41FGHH314 pKa = 7.58RR315 pKa = 11.84SHH317 pKa = 7.07ALANSFINPPLFSVPEE333 pKa = 3.87QLDD336 pKa = 3.82ANVFTSDD343 pKa = 3.35CVSNGRR349 pKa = 11.84YY350 pKa = 8.95FKK352 pKa = 11.13YY353 pKa = 10.89NEE355 pKa = 4.03FLRR358 pKa = 11.84DD359 pKa = 3.41SLGAIGGGANDD370 pKa = 4.44LLRR373 pKa = 11.84EE374 pKa = 3.95LMEE377 pKa = 3.73YY378 pKa = 10.31RR379 pKa = 11.84RR380 pKa = 11.84LGGSTSDD387 pKa = 3.18KK388 pKa = 10.97LNYY391 pKa = 9.49LAYY394 pKa = 10.37FGNKK398 pKa = 7.6SQCDD402 pKa = 3.6TLLGLDD408 pKa = 3.69TAPVYY413 pKa = 11.5SMFVSGVVRR422 pKa = 11.84EE423 pKa = 4.8PYY425 pKa = 10.12VHH427 pKa = 6.23MPYY430 pKa = 10.55SVLVKK435 pKa = 10.48KK436 pKa = 8.42FTYY439 pKa = 9.89NAQASGGTLDD449 pKa = 3.88KK450 pKa = 11.41QFGFKK455 pKa = 10.42NKK457 pKa = 10.22SDD459 pKa = 3.78VLLLNLVAYY468 pKa = 10.16RR469 pKa = 11.84LISTAWCIQKK479 pKa = 10.2RR480 pKa = 11.84SKK482 pKa = 10.58LKK484 pKa = 10.49PGIIWDD490 pKa = 3.68SASRR494 pKa = 11.84PKK496 pKa = 10.4LIKK499 pKa = 10.14IAGTGNKK506 pKa = 8.05MKK508 pKa = 10.36KK509 pKa = 9.78IMAGEE514 pKa = 4.11PCCRR518 pKa = 11.84VIAVGPLLEE527 pKa = 4.42SLLGYY532 pKa = 9.97SLYY535 pKa = 10.91LRR537 pKa = 11.84VSEE540 pKa = 5.13RR541 pKa = 11.84LQQFFKK547 pKa = 10.89EE548 pKa = 4.0KK549 pKa = 10.31GYY551 pKa = 11.13GVAIGCNRR559 pKa = 11.84MGKK562 pKa = 9.4DD563 pKa = 2.85WQNIQKK569 pKa = 9.33NFKK572 pKa = 8.69GARR575 pKa = 11.84HH576 pKa = 5.72IMVGDD581 pKa = 3.45YY582 pKa = 10.92SKK584 pKa = 11.52YY585 pKa = 10.39DD586 pKa = 3.18QTIPEE591 pKa = 4.12RR592 pKa = 11.84LMIFGIDD599 pKa = 3.97MILNMYY605 pKa = 8.49MPQDD609 pKa = 4.34EE610 pKa = 4.59YY611 pKa = 11.84SMNYY615 pKa = 9.02IDD617 pKa = 6.17NFRR620 pKa = 11.84TWFIDD625 pKa = 3.59NIVRR629 pKa = 11.84SVHH632 pKa = 5.71VVDD635 pKa = 5.28GKK637 pKa = 11.11LSARR641 pKa = 11.84AIGGMPSGTLWTSLLNSVINIVVISDD667 pKa = 3.4TCKK670 pKa = 10.45ALGIKK675 pKa = 9.77NYY677 pKa = 10.28KK678 pKa = 8.44PVVYY682 pKa = 10.11GDD684 pKa = 3.45DD685 pKa = 3.56HH686 pKa = 6.69MIIIYY691 pKa = 10.1DD692 pKa = 3.99EE693 pKa = 4.44IVDD696 pKa = 3.68SGEE699 pKa = 4.25FISAYY704 pKa = 7.91STYY707 pKa = 11.03VKK709 pKa = 10.38INFGMTLTTDD719 pKa = 3.35DD720 pKa = 4.46TYY722 pKa = 11.38ISGPEE727 pKa = 4.09CFYY730 pKa = 10.52VTYY733 pKa = 10.27KK734 pKa = 10.71RR735 pKa = 11.84PVYY738 pKa = 10.23NPKK741 pKa = 10.51ADD743 pKa = 3.71LQKK746 pKa = 9.53GTRR749 pKa = 11.84RR750 pKa = 11.84LRR752 pKa = 11.84PKK754 pKa = 10.08SWEE757 pKa = 3.87TSKK760 pKa = 9.95TPFTQFDD767 pKa = 3.68HH768 pKa = 6.71TKK770 pKa = 9.03GTTHH774 pKa = 6.47RR775 pKa = 11.84WSYY778 pKa = 9.02EE779 pKa = 3.25FRR781 pKa = 11.84GRR783 pKa = 11.84PKK785 pKa = 10.21FLQYY789 pKa = 11.01YY790 pKa = 8.1WLEE793 pKa = 3.97TGLAIRR799 pKa = 11.84PLRR802 pKa = 11.84EE803 pKa = 3.69SMVRR807 pKa = 11.84MLHH810 pKa = 6.52PEE812 pKa = 3.93EE813 pKa = 4.01EE814 pKa = 4.51VKK816 pKa = 10.66NVRR819 pKa = 11.84EE820 pKa = 3.98YY821 pKa = 9.65EE822 pKa = 4.27VLVISHH828 pKa = 7.32LYY830 pKa = 10.34DD831 pKa = 3.86NYY833 pKa = 11.1HH834 pKa = 5.99NAHH837 pKa = 5.2MRR839 pKa = 11.84NWAFHH844 pKa = 6.59LLYY847 pKa = 10.7DD848 pKa = 3.53IDD850 pKa = 3.89FMKK853 pKa = 10.42RR854 pKa = 11.84ARR856 pKa = 11.84MKK858 pKa = 10.79SADD861 pKa = 3.84LFFKK865 pKa = 10.8RR866 pKa = 11.84SDD868 pKa = 3.37RR869 pKa = 11.84GKK871 pKa = 10.92GYY873 pKa = 10.51YY874 pKa = 8.53EE875 pKa = 4.3HH876 pKa = 7.17NKK878 pKa = 9.84GPPSRR883 pKa = 11.84MWYY886 pKa = 9.84RR887 pKa = 11.84RR888 pKa = 11.84VDD890 pKa = 3.59YY891 pKa = 10.93LVDD894 pKa = 4.19LDD896 pKa = 4.25SCVGMQHH903 pKa = 6.54FVHH906 pKa = 5.85SWKK909 pKa = 10.75VLLAQMDD916 pKa = 4.26EE917 pKa = 4.75IISSEE922 pKa = 4.34HH923 pKa = 5.35EE924 pKa = 4.03CEE926 pKa = 4.37PYY928 pKa = 10.0QVRR931 pKa = 11.84HH932 pKa = 6.36LIKK935 pKa = 10.65NEE937 pKa = 3.24LSMRR941 pKa = 11.84RR942 pKa = 11.84ISTTRR947 pKa = 11.84ARR949 pKa = 11.84AWFEE953 pKa = 3.43AGIITRR959 pKa = 11.84SEE961 pKa = 4.06YY962 pKa = 11.15AQFLAAEE969 pKa = 4.64GTSKK973 pKa = 10.42QFEE976 pKa = 4.72SKK978 pKa = 10.43KK979 pKa = 10.8SPFNLFGLDD988 pKa = 3.29EE989 pKa = 4.37VGFNYY994 pKa = 10.31LFSLLGISNDD1004 pKa = 3.12KK1005 pKa = 10.18VHH1007 pKa = 6.68YY1008 pKa = 9.25STDD1011 pKa = 3.47YY1012 pKa = 11.35DD1013 pKa = 3.67RR1014 pKa = 11.84GPSRR1018 pKa = 11.84LKK1020 pKa = 10.42QVLATSIDD1028 pKa = 3.98LNGFASVVNFASKK1041 pKa = 9.77WLQSSDD1047 pKa = 3.43SPNLYY1052 pKa = 8.81RR1053 pKa = 11.84TGEE1056 pKa = 4.08PPP1058 pKa = 3.37
Molecular weight: 122.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1351 |
293 |
1058 |
675.5 |
78.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.625 ± 0.393 | 1.11 ± 0.04 |
5.996 ± 0.089 | 6.44 ± 1.27 |
5.922 ± 0.57 | 5.329 ± 0.877 |
1.925 ± 0.412 | 5.848 ± 0.292 |
8.142 ± 1.272 | 9.845 ± 0.18 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.813 ± 0.194 | 5.329 ± 0.216 |
3.183 ± 0.676 | 3.997 ± 0.67 |
5.922 ± 0.367 | 7.624 ± 0.209 |
4.071 ± 0.145 | 5.107 ± 0.463 |
1.258 ± 0.263 | 4.515 ± 1.129 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
